Figure S3.
PRC1 Has a Genome-wide-Role in PRC2 Recruitment and Polycomb Domain Formation at Target Sites in Mouse ESCs, Related to Figure 4
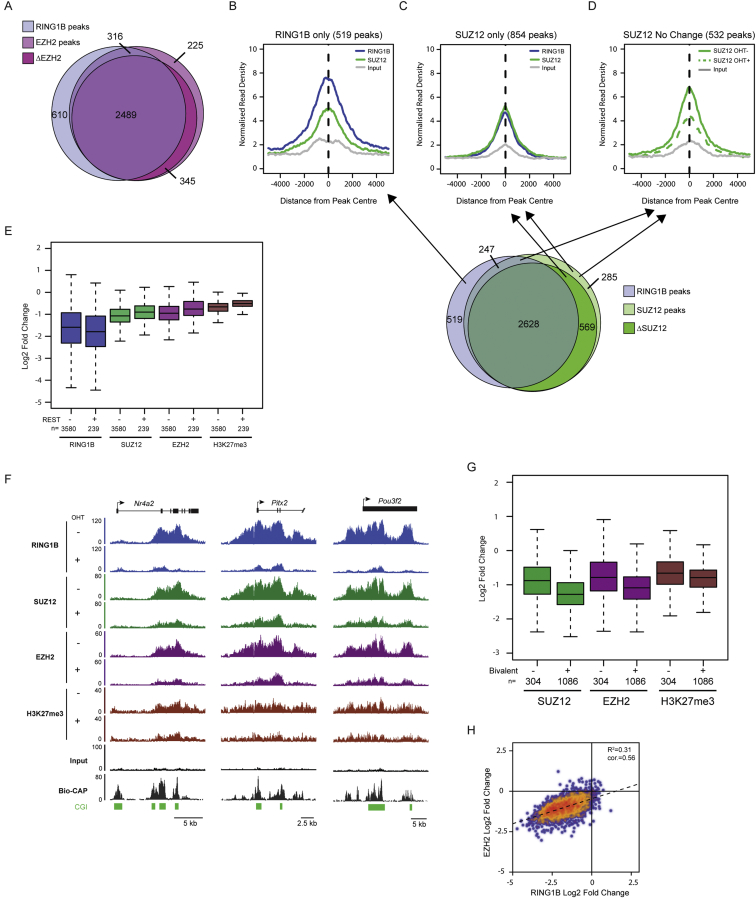
(A) A Venn diagram showing the overlap of RING1B (light blue) and EZH2 (light pink) peaks including a further segregation of EZH2-bound regions that show a greater than 1.5-fold change in EZH2 occupancy (ΔEZH2, dark pink) after RING1A/B deletion. The majority of EZH2-bound locations show changes in EZH2 signal following removal of RING1A/B, and most of the changes are restricted to sites associated with RING1B peaks.
(B) A metaplot of RING1B and SUZ12 ChIP-seq data at RING1B peaks that do not overlap with SUZ12 sites identified by peak calling. This indicates that these regions exhibit SUZ12 enrichment but are likely below the enrichment level required for peak detection.
(C) A metaplot of RING1B and SUZ12 ChIP-seq data at SUZ12 peaks that do not overlap with RING1B sites identified by peak calling. This indicates that these regions exhibit RING1B enrichment but are likely below the enrichment level required for peak detection.
(D) A metaplot of SUZ12 ChIP-seq data in the Ring1a−/−Ring1bfl/flb cells at SUZ12 peaks exhibiting a less than 1.5-fold reduction in SUZ12 signal following tamoxifen treatment. Importantly these sites still show reduction in SUZ12 signal suggesting that loss of RING1A/B affects PRC2 occupancy at most sites.
(E) A box and whisker plot indicating the Log2 fold change in polycomb factors at SUZ12-bound sites with and without REST following loss of RING1A/B. This indicates that loss of PRC2 occurs at REST-bound sites in the absence of PRC1.
(F) Snapshots of ChIP-seq traces for RING1B, SUZ12, EZH2 and H3K27me3 in the Ring1a−/−Ring1bfl/fl cells prior to (−OHT) and following 48 hr (+OHT) of tamoxifen treatment at sites previously reported to rely on the Meg3 long noncoding RNA for PRC2 targeting. In all cases we observe appreciable loss of PRC2 following RING1A/B deletion indicating that Meg3-dependent targeting is not sufficient to maintain normal levels of PRC2 at these sites.
(G) A box and whisker plot indicating the Log2 fold change in PRC2 factors and H3K27me3 at sites considered to be bivalent. Bivalent sites appear to have slightly larger fold changes in PRC2 occupancy following RING1A/B deletion.
(H) A scatter plot comparing the fold change of RING1B and EZH2 at EZH2 peaks. This indicates a clear correlation between the magnitude in RING1B and EZH2 change suggesting these changes may be mechanistically linked.