Figure 8.
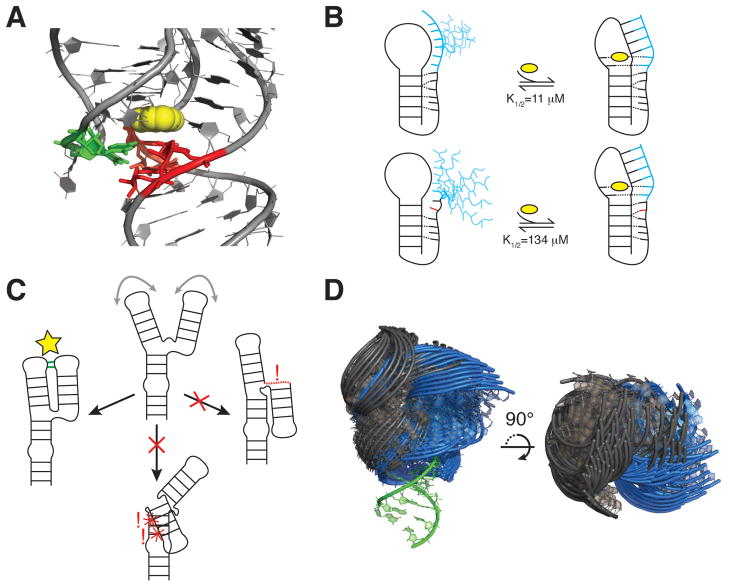
Interdependencies of CS across Tiers. (A) Aptamer domain of the add adenine riboswitch in complex with adenine (yellow) (PDB ID 1Y26). P1 stem base pairs shown to be unstable in the absence of ligand are shown in red (30), with J2/3 residues that provide stabilizing A-minor interactions shown in green. (B) Stacking interactions limit loop dynamics and pre-organize the 3’ tail for ligand binding and pseudoknot folding in the wild-type Bsu preQ1 riboswitch aptamer (top). An A-to-C mutation distal from the ligand binding pocket disrupts stacking, increasing dynamics and reducing ligand/riboswitch affinity (bottom) (176). The preQ1 ligand, 3’ tail, and mutation are shown in yellow, blue, and red, respectively. (C) Topological constraints preclude a three-way junction from forming two of three possible tertiary interactions. Right, the interaction is precluded due to connectivity. Bottom, the interaction is precluded due to sterics. (D) View of 50 most probable inter-helical conformations for two 1-nt bulge junctions with lower stem superimposed (green). The bulge of the blue junction is located two base-pairs below that of the grey junction. Most probable conformations were obtained from coarse-grained model simulations that include only steric and connectivity forces (178).