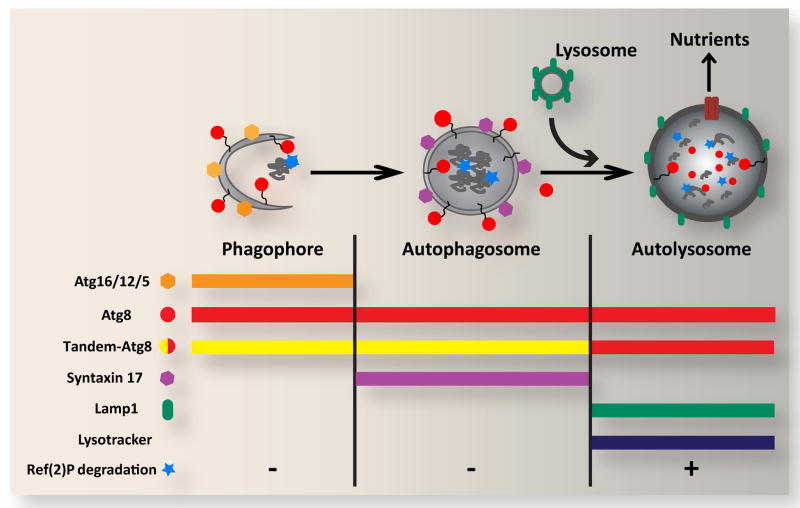
Figure 1. Overview of autophagic structures, markers and assays.
In response to cellular stress and other inductive signals, cytoplasm, protein aggregates and organelles are engulfed within double-membraned autophagosomes, which form through growth of the cup-shaped phagophore/isolation membrane. Fusion of the outer autophagosomal membrane with endosomes (not shown) and lysosomes results in exposure of the sequestered material to the hydrolytic lysosomal compartment. Degraded materials are ultimately exported to the cytoplasm as nutrients for energy and biosynthesis. Each of these steps can be identified and distinguished based on their specific constellation of associated proteins, as well as by pH and degradative properties. Atg5, Atg12 and Atg16 form a complex on the outer surface of the phagophore, and are removed upon its closure to form the autophagosome. Syntaxin 17 is recruited to the mature autophagosome, and is released upon fusion with the lysosome. In contrast, Atg8 is associated with the entire autophagic pathway from phagophore to autolysosome. Autophagosome-lysosome fusion can be monitored through loss of GFP fluorescence from the tandem GFP-mCherry-Atg8 marker. General lysosomal markers such as Lamp1 and LysoTracker can also be used to identify later autophagic vesicles. The degradative capacity of autophagy can be monitored through the cytoplasmic levels or specific cleavage of the autophagy substrate Ref(2)P.