Fig. 1.
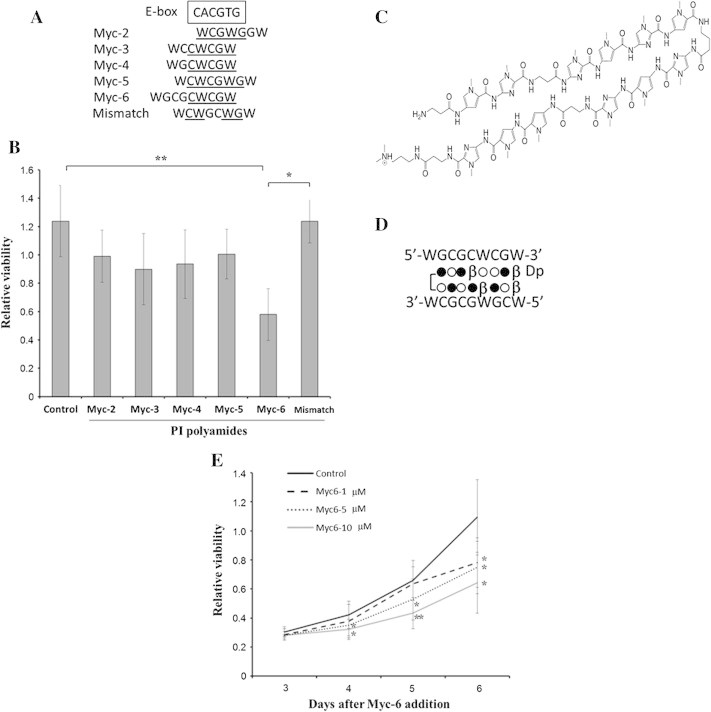
Myc-6 significantly represses cell viability of human osteosarcoma MG63 cells. (A) Sequence comparison between the putative targets of the indicated synthetic PI polyamides and the consensus E-box. Sequences corresponding to E-box are underlined. W indicates A or T. (B) WST-8 assay. MG63 cells were exposed to water (control), non-specific PI polyamide (mismatch) or with the indicated PI polyamides. Six days after the treatment, cell viability was examined by WST-8 assay. Data are expressed as the means ± SD. Differences were considered significant at p < 0.05. ∗p < 0.01;∗∗p < 0.001. (C) Chemical structure of Myc-6. (D) Schematic representation of DNA recognition by Myc-6. Open circle, filled circle, β and Dp indicate pyrrole, imidazole, β-alanine and dimethylpropanediamine, respectively. (E) Dose and time-dependency. Cell viability was measured by WST8 assay 3–6 days after addition of Myc-6. Differences between viability of cells treated with or without Myc-6 were considered significant at p < 0.05. ∗p < 0.05, ∗∗p < 0.01. Data were expressed as the means ± SD.
