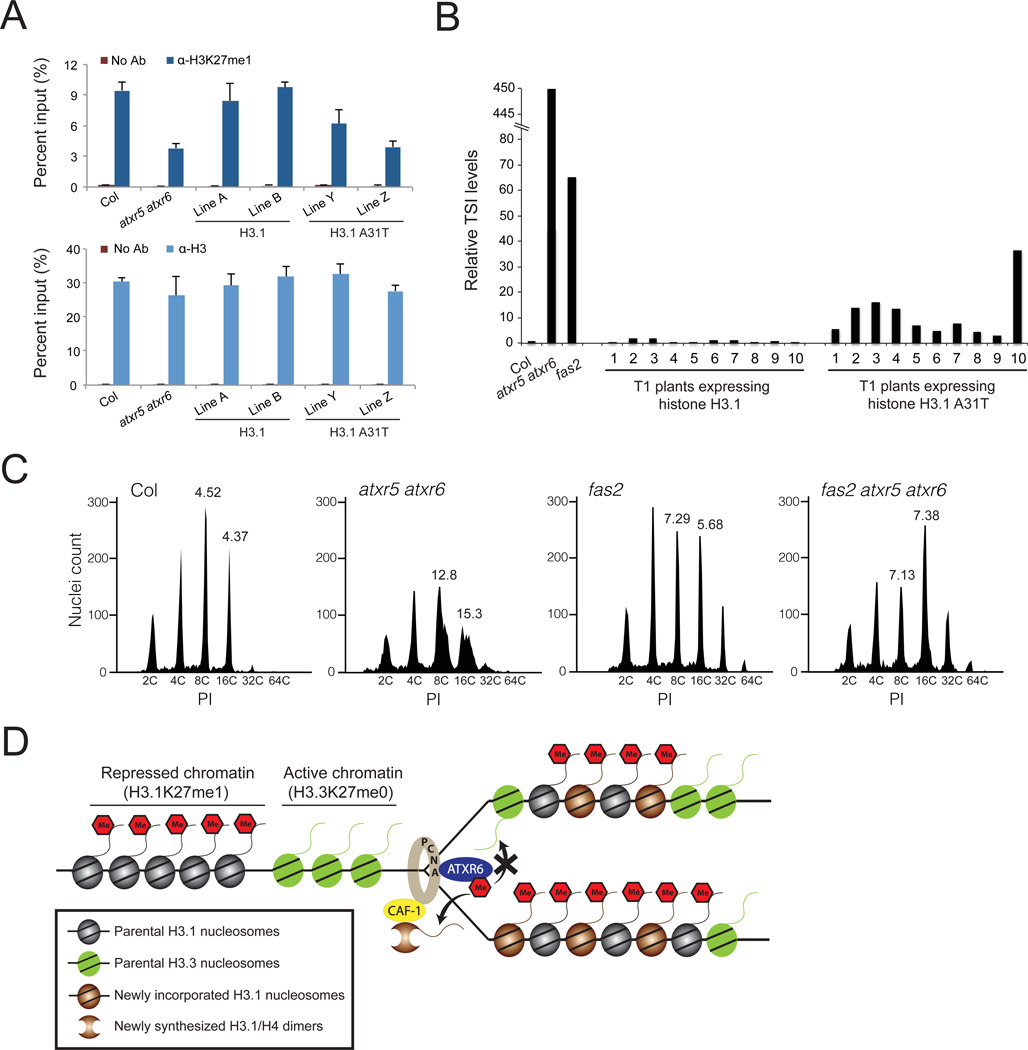
Figure 4. Thr-31 of H3.3 inhibits the activity of ATXR5/6 in vivo.
A. ChIP assays for H3K27me1 (upper panel) and H3 (lower panel) at TSI in transgenic T4 (homozygous) lines expressing H3.1 or H3.1 A31T. The average and standard deviation of three independent experiments is presented. ChIP for H3 serves as a control for nucleosome density. No Ab: no antibody control. B. RT-qPCR expression analysis of the repetitive element TSI in Col, atxr5 atxr6, fas2 and independent transgenic lines (first generation) expressing either wild-type plant H3.1 or the mutant H3.1 A31T. C. Flow cytometry profiles of Col, atxr5 atxr6, fas2 and fas2 atxr5 atxr6 leaf nuclei. The numbers below the peaks indicate the endoreduplication (ploidy) levels of the nuclei. The numbers above the 8C and 16C peaks correspond to the robust CV values (PI units that enclose the central 68% of nuclei) for those peaks. High robust CV values at 8C and 16C peaks characterize heterochromatic over-replication (4). PI: propidium iodide. D. Model for the role of ATXR5/6 during DNA replication in plants.