Fig. 5.
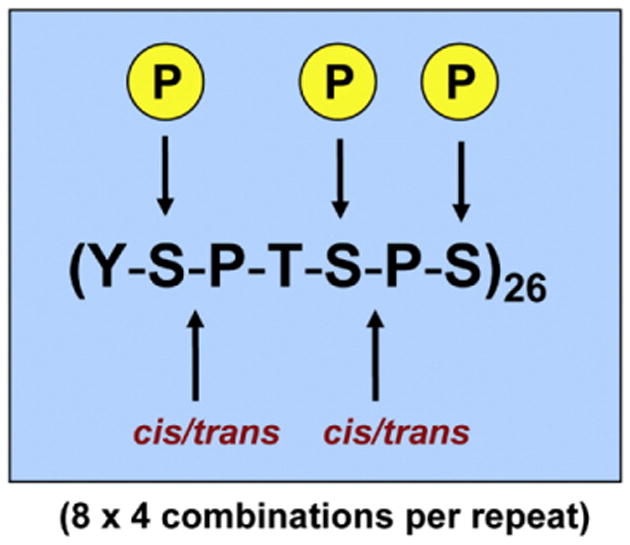
Simplified version of the CTD code. One repeat of the heptad sequence found at the carboxy-terminus of the Rpb1 subunit is shown. There are 26 repeats in yeast and 52 in humans. The Ser2, Ser5 and Ser7 residues can be phosphorylated in any combination (8 total), and the Ser2-Pro3 and Ser5-Pro6 bonds can exist in the cis or trans conformation (4 different combinations). Thus, there are at least 8 × 4 = 32 (×26 repeats) for a total of 832 potential configurations. Not shown are the potential phosphorylations at Tyr1, Thr4, as well as glycosylation at Thr4, and in humans, acetylation and methylation at degenerate Arg and Lys residues at position 7. See Egloff et al. [157] and other reviews cited in the text for details.
