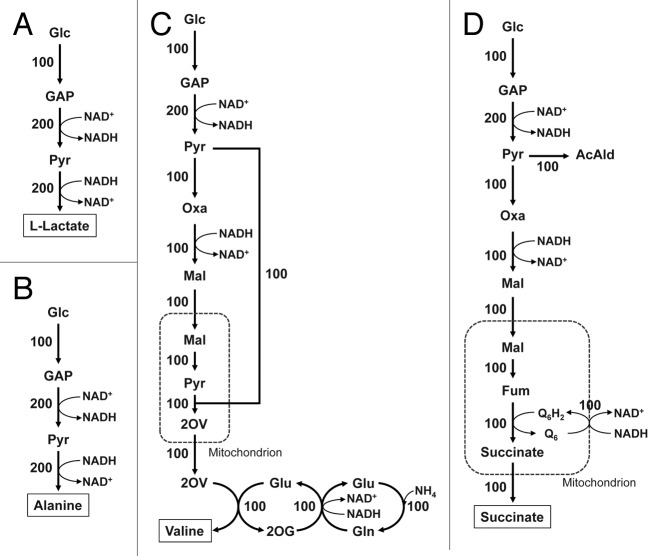
Figure 3. Simulated metabolic flux distributions during production of L-lactate (A), alanine (B), valine (C) and succinate (D) under anaerobic conditions in the recombinant strain incapable of both ethanol and glycerol biosynthesis. Metabolic simulation was performed using a genome-scale metabolic model of S. cerevisiae iND750 based on the concept of flux balance analysis; glucose and oxygen uptake rates assumed 1 and 0 mmol g dry cell−1 h−1, respectively, and the metabolic flux distributions at the metabolic steady-state were estimated as the biomass formation was maximized using linear programming. In these simulations, the fluxes for biosynthesis of the identified 20 metabolites, except for the target metabolite, as well as ethanol and glycerol biosynthesis were set to zero in advance. For simulation of L-lactate production (A), the heterologous reaction corresponding to LDH was added. In all cases, no biomass formation was achieved. The metabolic flux distributions normalized by the glucose uptake rate as 100 are shown. Glc, glucose; GAP, glyceraldehyde-3-phosphate; Pyr, pyruvate; Oxa, oxaloacetate; Mal, malate, Fum, fumarate; 2OV, 2-oxoisovalerate; AcAld, acetoaldehyde; Q6H2, ubiquinone (reduced form); Q6, ubiquinone (oxidized form).

An official website of the United States government
Here's how you know
Official websites use .gov
A
.gov website belongs to an official
government organization in the United States.
Secure .gov websites use HTTPS
A lock (
) or https:// means you've safely
connected to the .gov website. Share sensitive
information only on official, secure websites.