Abstract
The nuclear pore complex (NPC) is an enormous proteinaceous complex composed of multiple copies of about 30 different proteins called nucleoporins. In this study, we analyzed the composition of the NPC in the model organism Schizosaccharomyces pombe using strains in which individual nucleoporins were tagged with GFP. We identified 31 proteins as nucleoporins by their localization to the nuclear periphery. Gene disruption analysis in previous studies coupled with gene disruption analysis in the present study indicates that 15 of these nucleoporins are essential for vegetative cell growth and the other 16 nucleoporins are non-essential. Among the 16 non-essential nucleoporins, 11 are required for normal progression through meiosis and their disruption caused abnormal spore formation or poor spore viability. Based on fluorescence measurements of GFP-fused nucleoporins, we estimated the composition of the NPC in S. pombe and found that the organization of the S. pombe NPC is largely similar to that of other organisms; a single NPC was estimated as being 45.8–47.8 MDa in size. We also used fluorescence measurements of single NPCs and quantitative western blotting to analyze the composition of the Nup107-Nup160 subcomplex, which plays an indispensable role in NPC organization and function. Our analysis revealed low amounts of Nup107 and Nup131 and high amounts of Nup132 in the Nup107-Nup160 subcomplex, suggesting that the composition of this complex in S. pombe may differ from that in S. cerevisiae and humans. Comparative analysis of NPCs in various organisms will lead to a comprehensive understanding of the functional architecture of the NPC.
Keywords: nuclear pore complex, nucleoporins, Nup107-160 complex, nuclear envelope, fission yeast, fluorescence measurement
Introduction
The cell nucleus is enclosed by the nuclear envelope (NE), a double-membrane structure, which physically separates the nucleoplasm as a transcription space and the cytoplasm as a translation space, in eukaryotes. Molecules in the nucleus and the cytoplasm can be transported across the NE through the nuclear pores. The nuclear pore is an enormous protein complex, named the nuclear pore complex (NPC), that penetrates the NE. The molecular mass of the NPC is estimated to be ~125 MDa in vertebrate cells1 and ~50 MDa in yeast cells.2 The NPC is an 8-fold rotational symmetrical structure composed of multiple copies of approximately 30 different protein components called nucleoporins.2,3 A few transmembrane- and several scaffold-nucleoporins form the NPC-core structure that penetrates the nuclear membrane, and another group of nucleoporins form the NPC-peripheral layer which associates with the NPC-core structure. The core and peripheral layer are connected by adaptor nucleoporins. The NPC structure and most nucleoporins are conserved among eukaryotes.2-11
One of the subcomplexes forming the NPC-core structure is the Nup107-Nup160 subcomplex, composed of Nup160, Nup133, Nup107, Nup85, Nup96, Sec13, and Seh1, and depending on the species, additional nucleoporins Nup37, Nup43, and ELYS.12-15 Structural analysis of this complex has revealed that recombinant Nup107-Nup160 subcomplex proteins form a Y-shaped structure, and thus the complex is also referred as the Y-complex.16,17 The existence of the Y-complex is well conserved among eukaryotes.4,9,10,12,13,18-20 One of the important functions of the Nup107-Nup160 subcomplex is to assemble other nucleoporins or NPC subcomplexes into a complete and functional NPC after mitosis. In addition, the Nup107-Nup160 complex plays a role in kinetochore function.15,21-23 Thus, the Nup107-Nup160 complex is indispensable for NPC organization and chromosome segregation in eukaryotes.
The fission yeast Schizosaccharomyces pombe is an invaluable model experimental organism used for the elucidation of fundamental biological processes. The function and architecture of the NPC is also studied in S. pombe,19,24-34 but compared with budding yeast and humans in which NPC organization and function are intensively studied, knowledge of the S. pombe NPC is limited. In the present study, we generated S. pombe strains expressing individual GFP-tagged nucleoporins. In these strains, the genomic sequence coding the target nucleoporin was replaced with a GFP tagged nucleoporin sequence resulting in the GFP tagged nucleoporin being expressed under the regulation of its natural promoter as the sole endogenous protein. Using these strains, we confirmed the localization of 26 nucleoporins listed in the S. pombe database (http://www.pombase.org/) to the nuclear periphery. We also confirmed the localization of the predicted nucleoporin, Nup37, to the nuclear periphery. In addition we characterized 3 other S. pombe proteins as nucleoporins: Tts1, Ely5, and Amo1. The organization of the S. pombe NPC is largely similar to that of other organisms. Intriguingly, however, our fluorescence and biochemical analysis in S. pombe revealed some variation in the composition of the Nup107-Nup160 subcomplex among eukaryotes that suggests possible variation in the composition of the S. pombe Nup107-Nup160 subcomplex. In addition, we systematically disrupted the genes of the 9 nucleoporins that had not been individually characterized previously. Our results coupled with the previous reports indicate that 15 of the 31 proteins we identified as nucleoporins are essential for vegetative cell growth. Finally, disruption of the 16 non-essential nucleoporins revealed that 11 of these nucleoporins are essential for meiosis and normal spore formation and/or spore viability.
Results
Nucleoporins in S. pombe
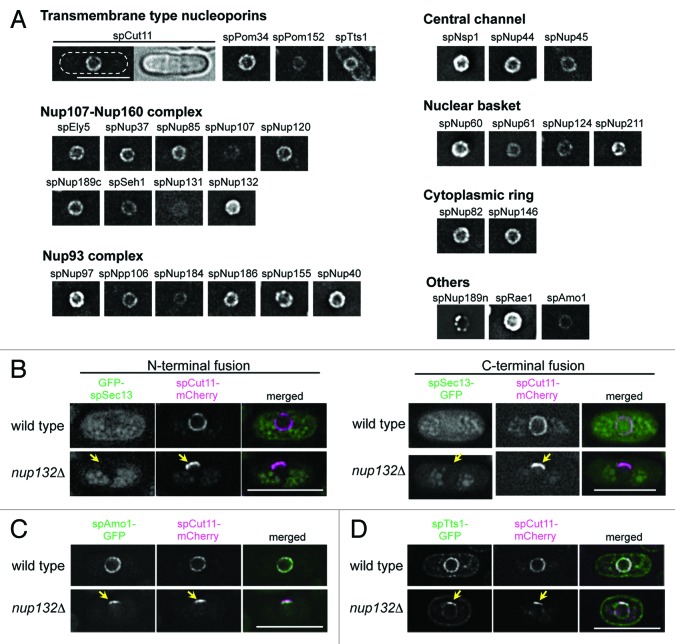
To comprehensively understand the nuclear pore complex (NPC) of S. pombe, we first selected 30 proteins as nucleoporins based on the S. pombe database (http://www.pombase.org/): 21 genes were selected based on experimental evidence and amino acid sequence similarities to proven nucleoporins,19,24-31,35 and the other 9 genes were selected based on amino acid sequence similarities alone (see Materials and Methods for details). Our previous study identified and named spEly5 and spNup37 (in this report we add the prefix “sp” for S. pombe) as nucleoporins in S. pombe,32 and during the course of the present study, Bilokapic and Schwartz independently characterized spEly5 and spNup37 as non-essential components of the S. pombe NPC.33 Thus, spEly5 and spNup37 are included in the group of “experimentally-identified nucleoporins” in Table S1. To examine whether these genes encode nucleoporins, individual target genes were replaced with GFP tagged sequences, as described in Materials and Methods, resulting in the GFP tagged nucleoporin being expressed under the regulation of its natural promoter as the sole endogenous protein. In these strains, since the gene is transcribed under its natural promoter, the GFP-fusion gene is expressed at physiological levels. Our strains expressing GFP-tagged nucleoporins were all viable, but four of them (spNup45-GFP, spNup184-GFP, GFP-spRae1, and spNup189n-GFP) showed growth deficiencies, and in one strain (spNup189n-GFP) localization of the GFP fluorescence was unusual; it showed clustered localization to the nuclear periphery with some bright spots (see “spNup189n” in Fig. 1A) and often showed additional dots inside the nucleus as well.
Figure 1.S. pombe nucleoporins fused to GFP. (A) Subcellular localization of GFP-tagged nucleoporins. The different images were obtained with the same acquisition times and processed in parallel. Deconvolved images are shown. The top left two panels show fluorescence (left) and bright field (right) images of an spCut11-GFP expressing cell. Nucleoporins were classified into seven groups according to localization in the NPC inferred from localization of the budding yeast orthologs. The scale bar represents 10 μm. (B-D) Localization of GFP-spSec13 (B), spAmo1-GFP (C), and spTts1-GFP (D) in wild type and cells lacking spNup132 (nup132Δ); in (B), GFP was fused with N-terminus (left) and C-terminus (right) of spSec13. spCut11-mCherry was simultaneously observed as a known nucleoporin marker. Yellow arrows indicate NPC-clustering regions in nup132Δ cells (lower panels). The scale bars represent 10 μm.
Fluorescence microscopy of these strains revealed that 29 proteins, all of our target genes except for spSec13, were located at the nuclear periphery with non-uniform dots characteristics of NPC localization (Fig. 1A), suggesting they are nucleoporins. Sec13 is conserved among eukaryotes and a component of the Nup107-Nup160 complex in humans and S. cerevisiae;14,16,36,37 spSec13 is also conserved in S. pombe.38 spSec13, however, did not localize to the nuclear periphery when fused with GFP either at its N- or C-termini (Fig. 1B). To further test if spSec13 is a structural component of the NPC in S. pombe, we examined its localization in cells lacking spNup132. In these cells, the NPCs make a cluster on one side of the nucleus,19 and this clustering of NPCs provides a unique opportunity to identify structural components of the NPC. In cells lacking spNup132, GFP-spSec13 did not co-localize with spCut11-mCherry (Fig. 1B): the nucleoporin spCut11 fused with mCherry at its C-terminus was used as a marker of the NPC. These results suggest that spSec13 may not be a structural component of the NPC in S. pombe.
In addition to the 29 proteins identified as nucleoporins, two other nuclear membrane proteins in the S. pombe database, spAmo1 and spTts1, were selected as candidate nucleoporins because they had amino acid sequence similarities to the nucleoporins hsNlp1 (also known as hCG1 and NUPL2)/scNup42 (also known as scRip1)39 and hsTMEM33/scPom33/scPer33,40 respectively. spAmo1-GFP localized to the nuclear periphery similarly to the other nucleoporins. spTts1-GFP localized to the nuclear envelope and endoplasmic reticulum, as reported previously.41 These results suggest that these proteins are associated with the NPC in S. pombe. To further affirm that spAmo1 and spTts1 are structural components of the NPC in S. pombe, we examined their localization in cells lacking spNup132 as described above.19 In these cells, spAmo1-GFP co-localized with the NPC marker spCut11-mCherry (Fig. 1C). spTts1-GFP also co-localized with spCut11-mCherry, although a fraction stayed on the NPC-free region of the NE (Fig. 1D). These results suggest that spAmo1 and spTts1 are nucleoporins.
We searched for novel nucleoporins in S. pombe using two different GFP fusion libraries, in which S. pombe genes are individually fused to the GFP gene,42,43 but failed to find any proteins with a localization characteristic of a nucleoporin. In the S. pombe genome database, at least 6 uncharacterized genes (SPAC18G6.13, SPAC17A2.07c, SPBC409.11, SPCC70.04c, SPCC1919.04, and SPBC25B2.08) are registered as genes encoding nuclear envelope proteins according to the genome wide protein localization study of S. pombe.44 However, they are S. pombe-specific genes and have no apparent orthologs in other eukaryotes. Thus, in this study we designate 31 proteins as nucleoporins in S. pombe based on their localization (Table S1). The domain organization of each nucleoporin in S. pombe is indicated in Figure S1.
Quantification of the nucleoporins based on fluorescence intensity
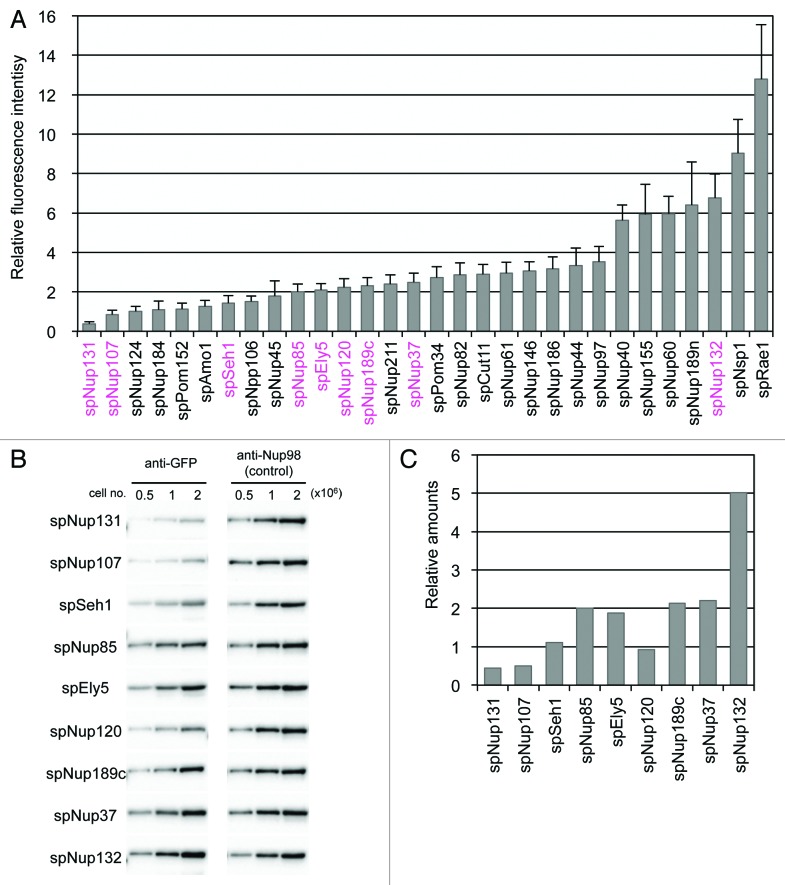
As the expression of GFP-tagged nucleoporins is under the control of the endogenous promoter, it is assumed that these proteins are generated in similar amounts as those of the native protein and that the composition of the NPC is the same in the strains generated in this study and wild-type cells. To determine the amount of the nucleoporins constituting the NPC, the fluorescence intensity of GFP in each strain was measured in living cells (see Materials and Methods for details): use of fluorescence intensity allows quantification of proteins present in the nuclear rim as opposed to quantifying the levels of proteins present throughout the cell. As expected, the fluorescence intensity of the different nucleoporins varied (Fig. 2A). Therefore, we estimated the relative values of the nucleoporins based on their fluorescence intensities. The fluorescence intensities and the relative values of the nucleoporins are listed in Table 1. Although spNup131, spNup107, and spSeh1 were also thought to form the Nup107-Nup160 subcomplex, they were separated from the other constituents because of their low fluorescence intensity (Fig. 2; Table 2). Notably, spNup131 had an extremely low fluorescence intensity. Because spNup131 has a paralog (spNup132), which shows relatively high fluorescence intensity, the small amount of spNup131 may be sufficient for its function in combination with spNup132 as an scNup133/hsNup133 homolog in S. pombe. Alternatively, it is possible that not all NPCs contain spNup131. The low fluorescence intensities of two other nucleoporins, spNup107 and spSeh1, may reflect a different Nup107-Nup160 subcomplex composition in S. pombe compared with that in S. cerevisiae and humans.
Figure 2. Quantification of GFP-tagged nucleoporins. (A) Fluorescence intensities of GFP-tagged nucleoporins. Fluorescence intensities of about 50 cells from each strain cultured at 26 °C were measured. Average values after background subtraction are shown in the bar graph. Error bars represent standard deviations. Components of the Nup107-Nup160 subcomplex are shown in magenta. (B) Quantitative western blot analysis of Nup107-Nup160 subcomplex nucleoporins. GFP-fused nucleoporins were fractioned by SDS-PAGE and detected by anti-GFP antibody (left). The membranes were stripped and reprobed to detect endogenous spNup189n as an internal control (right). The numbers above the images indicate cell numbers used to prepare the whole cell extract for each lane. (C) The relative amounts of Nup107-Nup160 subcomplex nucleoporins based on quantitative western blot analysis. Protein band intensities for 1 × 106 cells in (B) were measured for quantification. See Materials and Methods for detail.
Table 1. Nucleoporins in S. pombe.
| Name | ORF name | predicted MW | fluorescence intensity (unit) | Relative amount |
|---|---|---|---|---|
| spCut11 | SPAC1786.03 | 69.3 | 100.32 | 2.89 |
| spPom34 /Mug31 | SPAC1002.02 | 25.8 | 94.83 | 2.73 |
| spPom152 | SPBC29A10.07 | 139.5 | 39.31 | 1.13 |
| spTts1 | SPBC1539.04 | 31.5 | ND | ND |
| spEly5 | SPBC29A10.06c | 35.1 | 73.02 | 2.10 |
| spNup37 | SPAC4F10.18 | 42.7 | 86.60 | 2.49 |
| spNup85 | SPBC17G9.04c | 78 | 69.54 | 2.00 |
| spNup107 | SPBC428.01c | 93.2 | 29.75 | 0.86 |
| spNup120 | SPBC3B9.16c | 129.7 | 77.40 | 2.23 |
| spNup131 | SPBP35G2.06c | 131.4 | 13.32 | 0.38 |
| spNup132 | SPAC1805.04 | 132 | 235.30 | 6.77 |
| spNup189c | SPAC1486.05 | 94.6 | 80.09 | 2.30 |
| spSeh1 | SPAC15F9.02 | 38.5 | 49.13 | 1.41 |
| spNup97 /Mug87 | SPCC1620.11 | 97.5 | 122.35 | 3.52 |
| spNpp106 | SPCC1739.14 | 105.7 | 52.71 | 1.52 |
| spNup184 | SPAP27G11.10c | 176.9 | 37.90 | 1.09 |
| spNup186 | SPCC290.03c | 186.4 | 110.10 | 3.17 |
| spNup155 | SPAC890.06 | 147.6 | 206.04 | 5.93 |
| spNup40 | SPAC19E9.01c | 40.2 | 195.44 | 5.62 |
| spNsp1 | SPAC26A3.15c | 60.7 | 314.31 | 9.04 |
| spNup44 | SPBC19G7.15 | 44.4 | 115.94 | 3.33 |
| spNup45 | SPAC22G7.09c | 44.9 | 60.53 | 1.77 |
| spNup82 | SPBC13A2.02 | 90.6 | 99.41 | 2.86 |
| spNup146 | SPAC23D3.06c | 145.7 | 106.41 | 3.06 |
| spNup60 | SPCC285.13c | 80.2 | 207.13 | 5.96 |
| spNup61 | SPCC18B5.07c | 59.1 | 102.42 | 2.95 |
| spNup124 | SPAC30D11.04c | 123.9 | 35.09 | 1.01 |
| spNup211 | SPCC162.08c | 211.4 | 82.91 | 2.38 |
| spNup189n | SPAC1486.05 | 95 | 222.33 | 6.39 |
| spRae1 | SPBC16A3.05c | 38.6 | 444.89 | 12.80 |
| spAmo1 | SPBC15D4.10c | 51.7 | 43.34 | 1.25 |
Table 2. Essentiality of S. pombe nucleoporins.
| Name | ORF name | Individual analyses (references) | Comprehensive deletion analysis (Kim et al.)46 | This study | |
|---|---|---|---|---|---|
| spCut11 | SPAC1786.03 | e | (West et al.)26 | e | |
| spPom34 /Mug31 | SPAC1002.02 | n | n | ||
| spPom152 | SPBC29A10.07 | n | n | ||
| spTts1 | SPBC1539.04 | n | (Zhang et al.)41 | n | |
| spEly5 | SPBC29A10.06c | n | (Bilokapic and Schwartz)33 | n | n |
| spNup37 | SPAC4F10.18 | n | (Bilokapic and Schwartz)33 | n | n |
| spNup85 | SPBC17G9.04c | e | (Baï et al.; Chen et al.)19,29 | e | |
| spNup107 | SPBC428.01c | e | (Baï et al.; Chen et al.)19,29 | e | |
| spNup120 | SPBC3B9.16c | n | (Baï et al.)19 | n | |
| spNup131 | SPBP35G2.06c | n | (Baï et al.)19 | n | |
| spNup132 | SPAC1805.04 | n | (Baï et al.; Chen et al.)19,29 | n | |
| spNup189c | SPAC1486.05 | e | |||
| spSeh1 | SPAC15F9.02 | n | (Baï et al.; Chen et al.)19,29 | n | |
| spNup97 /Mug87 | SPCC1620.11 | e | (Cho et al.)30 | e | |
| spNpp106 | SPCC1739.14 | n | (Yoon et al.)25 | n | |
| spNup184 | SPAP27G11.10c | n | (Whalen et al.)28 | n | |
| spNup186 | SPCC290.03c | e | (Chen et al.)29 | e | |
| spNup155 | SPAC890.06 | n | e | ||
| spNup40 | SPAC19E9.01c | n | (Chen et al.)29 | n | |
| spNsp1 | SPAC26A3.15c | e | (Chen et al.)29 | e | |
| spNup44 | SPBC19G7.15 | e | (Chen et al.)29 | e | |
| spNup45 | SPAC22G7.09c | e | (Chen et al.)29 | e | |
| spNup82 | SPBC13A2.02 | n | e | ||
| spNup146 | SPAC23D3.06c | e | (Chen et al.)29 | e | |
| spNup60 | SPCC285.13c | n | n | ||
| spNup61 | SPCC18B5.07c | n | (Chen et al.)29 | n | |
| spNup124 | SPAC30D11.04c | n | (Balasundaram et al.)27 | n | |
| spNup211 | SPCC162.08c | e | (Chen et al.)29 | e | |
| spNup189n | SPAC1486.05 | e | |||
| spRae1 | SPBC16A3.05c | e | (Brown et al.)24 | e | |
| spAmo1 | SPBC15D4.10c | n | (Pardo and Nurse)48 | n | |
Essential nucleoporins are shown in bold according to individual analyses and this study. “e” and “n” represent “essential” and “non-essential,” respectively.
Quantification of Nup107-Nup160 subcomplex components based on western blotting
We also determined the approximate amounts of Nup107-Nup160 subcomplex proteins by quantitative western blot analysis (Fig. 2B); development of the membranes yielded single protein bands corresponding to each GFP-nucleoporin, suggesting that none of GFP-nucleoporins tested were degraded (Fig. S2). For these membranes, endogenous spNup189n was used as an internal control for normalization of the relative amounts of the GFP-nucleoporins; the specific spNup189n antibody clone 13C245 was used in the determination of the band intensity of endogenous spNup189n (Fig. 2B). The results of our western blot analysis agreed with the results of our fluorescence-based analysis (compare Fig. 2A and C): the S. pombe NPC contains low amounts of Nup107 and Nup131 and high amounts of Nup132 (Fig. 2A and C). This result suggests that the S. pombe Nup107-Nup160 subcomplex may have a different composition from that in S. cerevisiae and humans.
Estimation of nucleoporin amounts in a single NPC
Gross estimation by fluorescence and biochemistry suggested that the composition of the S. pombe NPC is different. Especially, the smaller amount of spNup107 is unique because Nup107 is an essential component for cell growth and is thought to be a constituent of a conserved core structure of the NPC in other organisms. The estimation by fluorescence was based on fluorescence intensities from a whole single nucleus and estimation by biochemistry was based on whole cell extracts. Consequently, the low amount of spNup107 in the S. pombe NPC may reflect diverse compositions of individual NPCs: some NPCs may harbor a higher number of spNup107 proteins, similar to the levels in the NPCs of S. cerevisiae and humans, while others may harbor lower numbers. To clarify the amounts of spNup107 present in individual NPCs, we measured fluorescence intensities of selected nucleoporins for single NPCs using highly inclined and laminated optical sheet (HILO) microscopy, which is a modification of total internal reflection fluorescence microscopy.46 The fluorescence of individual NPCs on the surface of the NE can be imaged using HILO microscopy (Fig. 3A). In this experiment, we used spNup85 as a standard and set the relative value of its fluorescence intensity as two. The number of spNup107-GFP foci in a single nucleus was similar to that of spNup85-GFP foci, but the fluorescence intensity per single NPC was lower than that of spNup85 (Fig. 3A and B). These results suggest that most or all S. pombe NPCs contain small amounts of spNup107. We also measured the amounts of spNup131-GFP and spNup132-GFP in single NPCs. The relative values of spNup131-GFP and spNup132-GFP were 0.4 and 4.8, respectively. These results suggest that the constituents of the Nup107-Nup160 subcomplex may be present in a different ratio in S. pombe than S. cerevisiae or humans.
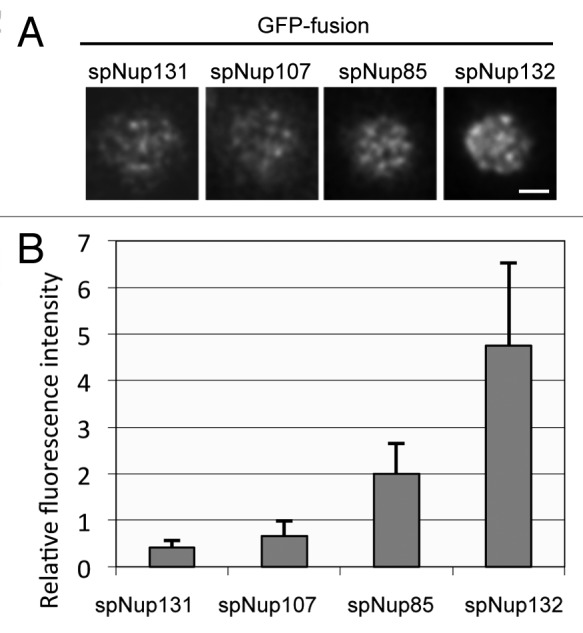
Figure 3. Fluorescence intensities of GFP-tagged nucleoporins measured by HILO microscopy. (A) HILO microscopy images of cells expressing spNup131-GFP, spNup107-GFP, spNup85-GFP, and spNup132-GFP. The nuclear region of a single cell is shown. The scale bar indicates 1 μm. (B) Fluorescence intensity of nucleoporins in single NPCs. Average and standard deviation are shown.
Our estimations, based on fluorescence intensities and biochemistry, suggest that some of the S. pombe nucleoporins, including spNup107, spSeh1, and spNup131, are present in the NPC in smaller amounts than in S. cerevisiae, although we cannot exclude the possibility that their intensities were underestimated for technical reasons. Based on the relative amounts obtained from the fluorescence intensities of the GFP-tagged nucleoporins, we estimate a molecular mass of a single S. pombe NPC as 45.8–47.8 MDa, similar to the value of ~50 MDa for S. cerevisiae.2,47
Essentialities of S. pombe nucleoporins
Previous studies on individual nucleoporins have reported 11 essential nucleoporins and 11 non-essential nucleoporins in S. pombe,19,24-30,41,48 and 6 additional non-essential nucleoporins were identified by comprehensive deletion analysis (see Table 2).49 Here we examined the 9 nucleoporins that had not been individually characterized previously (spEly5, spNup37, spNup189c, spNup155, spNup82, spNup60, spNup189n, spPom152, and spPom34) by gene disruption analysis using PCR-based gene targeting methods.50 We found that spNup155 and spNup82 were essential for vegetative cell growth and that spEly5, spNup37, spNup60, spPom152, and spPom34 were not essential. An alternative method was used for spNup189n and spNup189c because they are encoded by a single nup189+ gene transcribed as one transcript (HA and TH, unpublished result) and consequently endogenous nup189+ gene deletion results in deletion of both spNup189c and spNup189n. Therefore, nup189n was integrated into the lys1+ locus when analyzing spNup189c and nup189c was integrated into the aur1+ locus when analyzing spNup189n (see Materials and Methods for details). We found that cells were viable only when spNup189n and spNup189c were simultaneously expressed. Thus, we concluded that spNup189n and spNup189c were both essential for cell growth. Two of the nucleoporins, spNup155 and spNup82, reported to be nonessential by Kim et al.,49 were essential in our analysis (Table 2).
Taken together, gene disruption analysis in the previous studies combined with the gene disruption analysis of our study identified 15 nucleoporins essential for vegetative growth in S. pombe (Table 2). S. pombe homologs of essential nucleoporins in S. cerevisiae are also essential in cell growth in S. pombe (Table 3), suggesting that the NPC structure is largely similar among eukaryotes.
Table 3. Correspondence table of nucleoporins.
| S. pombe | S. cerevisiae | A. nidulans | H. sapiens | A. thaliana |
|---|---|---|---|---|
| spCut11 | scNdc1 | An-Ndc1 | hsNdc1 | At1g73240 |
| spPom34/Mug31 | scPom34 | An-Pom34 | - | - |
| spPom152 | scPom152 | An-Pom152 | - | - |
| spTts1 | scPom33 | AN6689.2 | hsTMEM33 | At3g02420 |
| scPer33 | ||||
| - | - | - | hsGp/Nup210 | atGp210 |
| - | - | - | hsPom121 | - |
| spEly5 | - | An-ELYS | hsELYS | atElys/HOS1 |
| spNup37 | - | An-Nup37 | hsNup37 | - |
| spNup85 | scNup85 | An-Nup85 | hsNup75/85 | atNup75 |
| spNup107 | scNup84 | An-Nup84 | hsNup107 | atNup107 |
| spNup120 | scNup120 | An-Nup120 | hsNup160 | atNup160 |
| spNup131 | scNup133 | An-Nup133 | hsNup133 | atNup133 |
| spNup132 | ||||
| spNup189c | scNup145c | SonBcNup96 | hsNup96 | atNup96/PRECOZ |
| spSeh1 | scSeh1 | An-Seh1 | hsSeh1/Sec13L | atSeh1 |
| ―* | scSec13 | An-Sec13 | hsSec13R | atSec13 |
| spNup97/Mug87 | scNic96 | An-Nic96 | hsNup93 | atNup93a |
| spNpp106 | atNup93b | |||
| spNup184 | scNup188 | An-Nup188 | hsNup188 | atNup188 |
| spNup186 | scNup192 | An-Nup192 | hsNup205 | atNup205 |
| spNup155 | scNup157 | An-Nup170 | hsNup155 | atNup155 |
| scNup170 | ||||
| spNup40 | scNup53 | - | hsNup35 (MP-44) | atNup35 |
| scNup59 | ||||
| spNsp1 | scNsp1 | An-Nsp1 | hsNup62 | atNup62 |
| spNup44 | scNup57 | An-Nup57 | hsNup54 | atNup54 |
| spNup45 | scNup49 | An-Nup49 | hsNup58 | atNup58 |
| - | - | - | hsNup45 | - |
| - | - | - | hsNup358 | - |
| spNup82 | scNup82 | An-Nup82 | hsNup88 | atNup88 |
| spNup146 | scNup159 | An-Nup159 | hsNup214 | atNup214 |
| spNup60 | scNup60 | - | - | - |
| spNup61 | scNup2 | An-Nup2 | hsNup50 | atNup50a |
| atNup50b | ||||
| spNup124 | scNup1 | - | hsNup153 | atNup136/Nup1 |
| spNup211 | scMlp1 | An-Mlp1 | hsTpr | atTpr/NUA |
| scMlp2 | ||||
| spNup189n | scNup100 | SonBnNup98 | hsNup98 | atNup98a |
| scNup116 | ||||
| atNup98b | ||||
| scNup145n | ||||
| spRae1 | scGle2 | SonAGle2 | hsRAE1 | atRAE1 |
| spAmo1 | scNup42/Rip1 | An-Nup42 | hsNlp1/hCG1/NUPL2 | - |
| - | - | - | hsALADIN | atALADIN |
| - | - | - | hsNup43 | atNup43 |
Essential and non-essential nucleoporins reported in S. pombe, S. cerevisiae, and A. nidulans are indicated in bold and italics, respectively. *Sec13 does not show nuclear periphery localization in S. pombe, thus it is not included in the “S. pombe” column and is represented as (-). Essentiality of AN6689.2 of A. nidulans is not clarified to date.
Functions of non-essential nucleoporins
We analyzed the 16 nucleoporins non-essential for vegetative cell growth. First, the growth of strains deleted for these nucleoporins was examined under various temperatures or in the presence of a DNA replication inhibitory drug, hydroxyurea (HU), or a microtubule-depolymerizing drug, thiabendazole (TBZ). In this assay, the colony size and colony number on agar plates were tested using a dilution series to compare growth rate and viability; strains deleted for Cds1 and Mph1 were used for positive controls to see the defects in the presence of HU and TBZ, respectively. The results are summarized in Table 4. Almost all strains could grow between 20 and 36 °C, but the strain lacking spNup120 showed severe temperature sensitivity: it could not grow at 36 °C, and formed small colonies even at 20 °C. The strain lacking spNpp106 or spAmo1 showed weak temperature sensitivity. Most strains showed growth inhibition on plates containing TBZ or HU, as judged by either small or fewer colonies. These results suggest that non-essential nucleoporins can be required to maintain cell viability when cell cycle progression is disturbed at DNA synthesis or mitosis.
Table 4. Function of non-essential nucleoporins for various growth conditions.
| genotypes | 26 °C | 30 °C | 36 °C | 20 μg/ml TBZ | 10 mM HU |
|---|---|---|---|---|---|
| wild type | ++++ | ++++ | ++++ | +++ | +++ |
| ∆pom34 | ++++ | ++++ | ++++ | +++ | +++ |
| ∆pom152 | ++++ | ++++ | ++++ | +++S | +++ |
| ∆ely5 | ++++ | ++++ | ++++ | ++ | + |
| ∆nup37 | ++++ | ++++ | ++++ | +++ | +++ |
| ∆nup120 | +++S | ++SS | - | - | - |
| ∆seh1 | ++++ | ++++ | ++++ | +++ | +++ |
| ∆nup131 | ++++ | ++++ | ++++ | +++ | +++ |
| ∆nup132 | ++++ | ++++ | ++++ | +SS | +S |
| ∆nup131 ∆nup132 | ++++ | ++++ | ++++ | +SSS | +S |
| ∆nup40 | ++++ | ++++ | ++++ | +++ | +++ |
| ∆nup184 | ++++ | ++++ | ++++ | +++ | +++ |
| ∆npp106 | ++++ | ++++S | +++SS | + | +++ |
| ∆nup60 | ++++ | ++++ | ++++ | +++ | ++SS |
| ∆nup61 | ++++ | ++++ | ++++ | +++ | +++ |
| ∆nup124 | ++++ | ++++ | ++++ | ++SS | ++SS |
| ∆amo1 | ++++S | +++S | +++S | +++ | +++ |
| ∆mph1 | ++++ | ++++ | ++++ | - | ++S |
| ∆cds1 | ++++ | ++++ | ++++ | +++ | - |
Each cell strain was cultured to midlog phase in liquid YES. 5-fold serial dilutions of each transformant were spotted onto YES plates with or without HU or TBZ. Viable colony number is indicated by -, +, ++, +++, and ++++: “-” indicates no colony formation. Colony size (growth rate) is indicated by S, for small; SS, for smaller; SSS, for smallest colonies. Δmph1 and Δcds1strains were used as positive controls sensitive to TBZ and HU, respectively. For ∆nup120 strains, TBZ sensitivity and HU sensitivity were assayed at 26 °C.
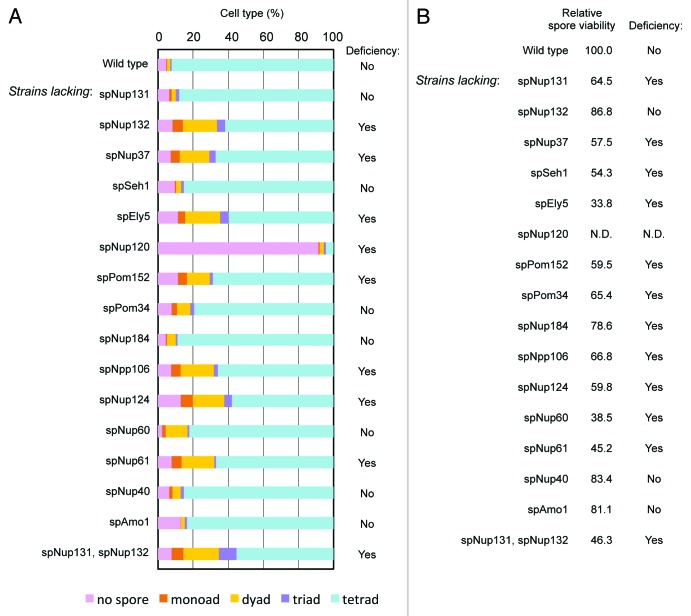
Next we examined meiotic functions of non-essential nucleoporins. In order to check meiotic phenotypes, deletion strains were induced to undergo meiosis, and spore formation and viability were examined. The wild type cell usually forms four spores following meiosis in S. pombe; however, in strains deleted for spNup132, spNup37, spEly5, spNup120, spPom152, spNpp106, spNup124, or spNup61 a fraction of the cells formed less than four spores (Fig. 4A). Furthermore, spore viability was low in strains lacking spNup131, spNup37, spSeh1, spEly5, spPom152, spPom34, spNpp106, spNup124, spNup60, or spNup61 (Fig. 4B); in cells lacking spNup120, spore viability was not determined because spore formation was severely defective. This result suggests an unidentified function of 11 non-essential nucleoporins in spore formation and/or spore viability in S. pombe.
Figure 4. Effect of gene disruption of non-essential nucleoporins on spore formation. (A) Spore numbers in wild type and nucleoporin mutants. Cells were cultured on sporulation medium for two days, and zygotic cells containing spores were differentially scored. “Yes” and “No” indicate cell strains with a deficiency and no deficiency in spore formation, respectively. (B) The numbers indicate the spore viability of wild type and nucleoporin mutants. Cells forming spores were treated with β-glucuronidase to digest vegetative cells, and the resistant spores were counted and spread on growth media. After colony formation, colony numbers were counted and spore viability was calculated. The value was normalized with that of the wild type, which was given a value of 100. “Yes” and “No” indicate cell strains with a deficiency and no deficiency in spore viability, respectively.
Discussion
The S. pombe NPC consists of evolutionary conserved nucleoporins: it includes an ELYS homolog and almost the same set of essential components as metazoans. Indeed, spEly5 and spNup37, which are nucleoporins lacking in the S. cerevisiae NPC, were recently reported to participate in the assembly of the S. pombe Nup107-Nup160 subcomplex and the crystal structure of spNup37 in the complex has been recently solved.33,34 This allows S. pombe to be used as a model organism for studies of the NPC.
Organization and specific features of the S. pombe NPC
We constructed a series of strains in S. pombe in which nucleoporin genes and genes similar to known nucleoporins were tagged with GFP. Fluorescence microscopy of the strains identified 31 nucleoporins (listed in Table 1) defined by their localization to the nuclear periphery. In this study, in addition to verifying 27 nucleoporins listed in the S. pombe database (http://www.pombase.org/) and the predicted nucleoporin spNup37 as nucleoporins, we characterized Tts1, Ely5, and Amo1 as nucleoporins. spNup37 and spEly5 have also been characterized as nucleoporins in other studies.32-34 Higher-order structures of nucleoporins are conserved, such as α-solenoid, β-propeller, and unstructured FG repeats, rather than amino acid sequences, consequently, there is poor similarity in amino acid sequence of S. pombe nucleoporins to human and budding yeast nucleoporins.
The organization of the NPC has been intensively studied in humans and budding yeast, and more recently in the nematode Caenorhabditis elegans,4 the filamentous fungus Aspergillus nidulans,6,20 the plant Arabidopsis thaliana,10 the divergent eukaryote Trypanosoma brucei,9 and the ciliate Tetrahymena thermophile.8,51 These studies have revealed that while there are species-specific differences in the NPC, the organization of the NPC is generally similar in the eukaryotes studied so far. This is also true of the S. pombe NPC. One of the features of the S. pombe NPC is that it includes spEly5/hsELYS and spNup37/hsNup37.20,32-34 These nucleoporins were originally identified as components of the Nup107-Nup160 subcomplex and thought to be included only in higher eukaryotic NPCs, as they are not found in the budding yeast. However, the ortholog of hsELYS and hsNup37 has been identified in the filamentous fungus Aspergillus nidulans,20 and now, in this study and others,32-34 in S. pombe. However, like S. cerevisiae and A. nidulans the S. pombe NPC lacks orthologs for hsNup45, hsNup358, hsALADIN and hsNup43. The unique characteristics of the S. pombe NPC likely reflect S. pombe-specific nuclear dynamics and/or life cycle.
Composition of the Nup107-Nup160 complex
We estimated the amount of endogenous nucleoporin by biochemistry and fluorescence microscopy of strains expressing physiological amounts of GFP-tagged nucleoporins. Our analysis suggests a unique organization of Nup107-Nup160 subcomplex in S. pombe. In S. cerevisiae and humans, each component of the Nup107-Nup160 subcomplex is thought to exist in equal amounts: one molecule of each nucleoporin is assembled into the Nup107-Nup160 subcomplex of the NPC, with two Nup107-Nup160 subcomplexes included in one NPC. In S. pombe, however, the relative ratios of spNup107 and spNup131 are lower compared with the other components, and the ratio of spNup132 is higher. Another feature in the S. pombe Nup107-Nup160 subcomplex is that spSec13 is lacking: GFP-spSec13 did not show localization to the nuclear periphery. Consistent with this result, biochemical isolation of the Nup107-Nup160 subcomplex did not identify spSec13.19 These data suggest that spSec13 is not associated with the Nup107-Nup160 subcomplex in S. pombe. It is possible that some other, unidentified protein, may substitute for Sec13 in the S. pombe Nup107-Nup160 subcomplex, but our analysis indicates that the organization of the S. pombe Nup107-Nup160 subcomplex may be different. Although we cannot exclude the possibility of artificial effects of GFP fusion and technical limitations of fluorescence intensity measurement, the unique organization of the S. pombe Nup107-Nup160 subcomplex suggests an evolutional diversity of the NPC and/or a species-specific function of the NPC in this organism. It will be important to follow the results of future proteomic and other parallel studies to see if they also support a potential difference in the stoichiometry of the S. pombe nuclear pore.
Functions of non-essential nucleoporins
While about half of the S. pombe nucleoporins are not essential for vegetative cell growth under normal laboratory conditions, they may be essential under certain circumstances. Strains deleted for non-essential nucleoporins enabled us to examine the effect of nucleoporin disruption on cell growth in the presence of a DNA replication inhibitory drug, HU, or a microtubule-depolymerizing drug, TBZ. In the presence of HU or TBZ, the non-essential nucleoporins spPom152, spEly5, spNup120, spNup132, spNpp106, spNup60, and spNup124 were required for cell growth, suggesting they have a role in chromosome function. An ELYS mutant caused decreased interaction of Mcm2, a protein required for DNA replication, with chromatin in zebrafish,52 and ELYS is required for genome stability in intestinal epithelium progenitor cells in the mouse,53 also suggesting a chromatin-related function for this nucleoporin. A role of NPCs in chromosome function has also been reported for many eukaryotes (for a review see refs. 54, 55). Further analysis is required to clarify whether the defects observed in the strains deleted for spPom152, spEly5, spNup120, spNup132, spNpp106, spNup60, or spNup124 are dependent on the nucleocytoplasmic transport function of the NPCs, a transport-independent function of the NPC, or an NPC-independent function of specific nucleoporin molecules.
In addition, the effect of non-essential nucleoporin gene disruption on the meiotic process was analyzed. Interestingly, most non-essential nucleoporins (11 out of 16) were required for meiosis and spore formation or spore viability. In particular, spNup60, spNup61, and spNup124, whose budding yeast orthologs localize at nucleoplasmic side of the NPCs,2 were all required for this processes in S. pombe. This result suggests important functions for nucleoplasmic components of the NPC in meiosis and spore formation. Consistent with this concept, the spNup61 ortholog is highly expressed in rat testis, suggesting a meiotic or developmental role.56 In A. thaliana, the mutant of the spNup124 ortholog, atNup136, displays early flowering and low fertility, indicating a function in plant development.10
Curiously, the strain lacking spNup131, which is thought to be a paralog of spNup132, showed low spore viability, but it did not show a defect in cell growth in the presence of HU or TBZ. The strain lacking spNup132, in contrast to the strain lacking spNup131, had a defect in spore formation but normal spore viability, and exhibited growth defects in the presence of both HU and TBZ. S. pombe is the only eukaryote that has two orthologs (spNup131 and spNup132) for hsNup133, and our analysis suggests they have distinct roles.
Meiosis and spore formation in S. pombe are thought to be an example of gametogenesis or cell differentiation that involves regulation of nutrition sensing, signal transduction and expression of specific transcription factors.57 The orthologs of nucleoporins required for meiosis and spore formation in S. pombe are also involved in cell differentiation in other eukaryotes. Nup133 (spNup131/spNup132 ortholog) is reported to have a role in neural cell differentiation in the mouse embryo;58 ELYS/Flo is required for progenitor cell proliferation in certain tissues in the zebrafish;54,59 Seh1 function is essential in fly oogenesis;60,61 the membrane-integrated nucleoporins, spPom34 and spPom152, are required for meiosis and spore formation in S. pombe; gp210/Nup210, a membrane-integrated nucleoporin in mammals, is expressed during oogenesis and is essential for myogenic and neuronal differentiation in the mouse;62-64 and gp210 is required for embryogenesis in Arabidopsis thaliana.65 On one hand, the cell differentiation process may require nucleocytoplasmic transport of specific molecules, including transcription factors. On the other hand, it is reported that the composition of the NPC may be altered during cell differentiation.52,64 A recent large-scale proteomics study of various types of human cells revealed varied composition of the NPC.66 Thus, the abnormal meiosis and spore formation observed in mutant S. pombe strains may be caused by failure of nucleocytoplasmic transport or by transport-independent functions of the NPC.
Investigation of nucleoporin function in S. pombe meiosis and spore formation may shed light on developmental roles of nucleoporins and provide information to help elucidate NPC and nucleoporin function not only in nucleocytoplasmic transport but also in cell differentiation in other eukaryotes.
Materials and Methods
Cell culture
YES medium was used as routine culture medium, and ME was used as sporulation medium. For measurement of fluorescence intensity of GFP-fused nucleoporins, strains were precultured in YES at 26 °C and then transferred to EMM2 medium at 26 °C.67 For subcellular localization analysis in cells lacking spNup132, cells were cultured on YES plate medium at 30 °C. To prepare drug-containing media, stock solutions of hydroxyurea (Sigma), 1 M in sterile water, and thiabendazole (Sigma), 20 mg/mL in dimethyl sulfoxide, were added to YES plates to the indicated final concentrations. Sporulation was induced at 26 °C.
Identification of nucleoporin genes in S. pombe
Candidates S. pombe nucleoporin genes were searched for in the S. pombe genome database (pombase, http://www.pombase.org/). We found 21 genes (genes encoding spCut11, spNup85, spNup107, spNup120, spNup131, spNup132, spNup189c, spSeh1, spNup97/Mug87, spNpp106, spNup186, spNup40, spNsp1, spNup44, spNup45, spNup146, spNup61, spNup124, spNup211, spNup189n, and spRae1) that were experimentally characterized as nucleoporins, and 9 genes (genes encoding spEly5, spNup37, spPom34/Mug31, spPom152, spNup184, spNup155, spNup82, spNup60, and spSec13) that were not experimentally characterized as nucleoporins but which share homologies with known nucleoporins in other eukaryotes. In addition, spAmo1 and spTts1 were identified as candidate nucleoporins because of homologies with known nucleoporins and their localization to the nuclear periphery.41,48
Generation of cell strains expressing fluorescent protein-tagged nucleoporins
We generated 31 cell strains, each with a different nucleoporin tagged with GFP, using AY160–14D (h90 ade6–216 ura4-D18 leu1–32 lys1–131) as a host strain. Sixteen nucleoporins (spCut11, spNup40, spNup44, spNup45, spNup61, spNup82, spNup85, spNup107, spNup120, spNup124, spNup131, spNup132, spNup146, spNup189c, spPom152, and spSeh1) were tagged with GFP-HA at their C-termini in a previous study.43 In this study, two nucleoporins (spEly5 and spTts1) were tagged with GFP-HA at their C-termini by a two-step PCR method described previously:43,68 these 18 GFP-HA tagged nucleoporins are denoted “-GFP” in the text. Ten nucleoporins (spNpp106, spNup155, spNup184, spNup186, spNup211, spAmo1, spNup37, spNup60, spNup189n, and spPom34) were tagged with GFP alone at their C-termini. spNup97 and spNsp1 were tagged with GFP at their N-termini: tagging these nucleoporins at the C-terminus was unsuccessful. In addition, spNup40, spNup124, spNup131, and spNup132 were also tagged with GFP at their N-termini: the fluorescence intensities of the C-terminus GFP-tagged proteins were weak and tagging with GFP at their termini N-termini increased the fluorescence signal. For spNup189n, we inserted an additional copy of the promoter sequence of spNup189 into the 3′ end of the marker module to ensure the physiological expression of downstream spNup189c. The GFP fusion constructs for each nucleoporin are summarized in Table S1. The pFA6a-mCherry-hphMX6 plasmid was used for tagging spCut11 with mCherry using the same PCR-based method as for tagging with GFP.69
Gene disruption
Gene disruption was performed with an ura4+ marker using PCR-based gene targeting methods.50 A diploid strain (h+/h- ade6-210/ade6-216 ura4-D18/ura4-D18 leu1-32/leu1-32 lys1-131/lys1-131) was used as the host strain. Diploid cells heterozygously deleted for a nucleoporin gene were induced to sporulate, and tetrad spores were dissected. Four viable spores in a tetrad set indicated that the nucleoporin gene was dispensable; two viable spores in a tetrad set indicated that the gene was essential.
To examine the importance of spNup189n and spNup189c in vegetative growth, a single entire ORF of the nup189+ gene was deleted in the diploid host strain, and then either nup189n or nup189c, or both, was integrated at the lys1+ or aur1+ locus. The spores generated in the diploid cell were dissected by micromanipulation and examined for cell growth. All of the haploid progeny were examined for ectopically-integrated spNup189n or spNup189c by checking their marker genes.
Image acquisition of GFP-nucleoporins by fluorescence microscopy
Living cells were mounted between coverslips.70 A DeltaVision microscope system (Applied Precision, Inc) based on an Olympus wide-field fluorescence microscope IX70 (Olympus Corp) was used for imaging. Images were obtained using an interline CoolSNAP HQ2 CCD camera (Photometrics, Tucson, USA) as an image detector through an oil-immersion objective lens (PlanApo 60×, NA, 1.4, Olympus). For measurement of fluorescence intensities, an oil-immersion objective lens (UApo 40×, NA, 0.65–1.35, Olympus) was used at NA = 0.65 (see below). Z-stack images were obtained at 0.4 μm interval for 8 Z-steps, and subjected to deconvolution to improve FM images by removing out-of-focus images.71
Estimation of nucleoporin amount based on fluorescence intensity
For measurement of fluorescence intensities, images of the cells expressing each GFP-nucleoporin were taken using a DeltaVision system as described above, using an oil-immersion objective lens (UApo 40×, NA, 0.65–1.35, Olympus) at NA = 0.65; simultaneously, images of the cell expressing spNup82-GFP were also taken as a control. The best-focused image of the eight optical sections was selected for quantification. The total fluorescence intensity of each nucleus was calculated as the sum of the intensities located in the nuclear region on the raw image using the SoftWoRx software equipped with the DeltaVision microscope system. The intensity values were normalized with that of spNup82-GFP. Fluorescence intensities of about 50 cells were measured in each strain, and average values were calculated after background subtraction. spNup85-GFP fluorescence intensity was set to two in calculating relative fluorescence intensities.
Western blot analysis
For quantitative western blot analysis, yeast whole cell extracts were prepared as follows. Yeast cells were cultured to log phase and incubated with 1 mM PMSF in the culture medium for 10 min. Five × 107 cells were then harvested and resuspended in 800 μl ice-cold water. The cell suspension was mixed with 150 μl of 1.85 N NaOH and incubated for 10 min on ice. Then, 150 μl of 55% (w/v) trichloroacetic acid was added and the mixture was incubated for 10 min on ice. The cells were centrifuged at 10 000 rpm for 2 min at 4 °C, and the pellet was washed with cold acetone. The pellet was then resuspended in sample buffer (200 mM Tris [pH 6.8], 8 M urea, 5% SDS, 0.1 mM EDTA, 100 mM DTT) and used as whole cell extract. Volume equivalents for cell number were calculated, and three different amounts of extract were applied and run by 8% SDS-PAGE. Proteins were transferred to a PVDF membrane in 25 mM Tris, 192 mM glycine, 10% methanol by wet transfer. The membrane was blocked with 5% skim milk in PBS containing 0.1% Tween20 and incubated with a rabbit polyclonal anti-GFP antibody (1:4000; Rockland Immunochemicals Inc) for 2 h at room temperature. The membrane was washed with PBS three times, and then incubated with secondary antibodies (1:2000) for 2 h at room temperature: HRP-conjugated goat anti rabbit IgG (GE Healthcare) was used as the secondary antibody. To ensure equal loading, the same membranes were reprobed for endogenous spNup189n using a mouse monoclonal anti-Nup98 antibody (13C2; 1:10 000)45 and HRP-conjugated goat anti-mouse IgG as described above. For quantitative western blot analysis, the band intensity of spNup189n was used for normalization. Protein bands were detected by chemiluminescence using ChemiDoc MP imaging system (Bio-Rad), and band intensities were measured using Image Lab software (Bio-Rad). Band intensity of spNup85 was set to two in calculating relative band intensities.
Measurement of nucleoporin amounts per a single NPC
Living cells were mounted in a glass-bottom culture dish coated with 2 mg/mL concanavalin A70 and were imaged with HILO microscopy as previously described.46 A solid-state laser (488 nm, 20 mW; Sapphire 488-20-OPS, Coherent) and an inverted microscope (IX-81, Olympus) equipped with an oil-immersion objective lens (PlanAPO 100×, NA 1.45, Olympus) were used. Images were captured with a back-thinned electron multiplying charge-coupled device camera (C9100-13; Hamamatsu Photonics) at a frame rate of 30 frames per second, and recorded on a digital hard-disk recorder (AQUACOSMOS, Hamamatsu Photonics). Fluorescence intensities were measured using Image J software.
Supplementary Material
Disclosure of Potential Conflicts of Interest
No potential conflict of interest was disclosed.
Acknowledgments
We thank Shelly Sazer and Paul Nurse for mph1 and cds1 strains and Aki Hayashi, Chie Mori, Miho Yamane, and Kasumi Okamasa for their technical assistances. We also thank DB Alexander for critical reading of the manuscript. This work was supported by grants from the Japan Science and Technology Agency (to T.H.) and the Ministry of Education, Culture, Sports, Science, and Technology of Japan (to H.A., Y.C., M.T., M.I., Y.H., and T.H.), and by a grant (H.A., Y.C., and M.I.) and a fellowship (H.-J. Y.) from the Japan Society for the Promotion of Science.
Glossary
Abbreviations:
- FG repeat
Phe-Gly repeat
- HILO microscopy
highly inclined and laminated optical sheet microscopy
- HU
hydroxyurea
- NE
nuclear envelope
- NPC
nuclear pore complex
- TBZ
thiabendazole
References
- 1.Reichelt R, Holzenburg A, Buhle EL, Jr., Jarnik M, Engel A, Aebi U. Correlation between structure and mass distribution of the nuclear pore complex and of distinct pore complex components. J Cell Biol. 1990;110:883–94. doi: 10.1083/jcb.110.4.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–51. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol. 2002;158:915–27. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galy V, Mattaj IW, Askjaer P. Caenorhabditis elegans nucleoporins Nup93 and Nup205 determine the limit of nuclear pore complex size exclusion in vivo. Mol Biol Cell. 2003;14:5104–15. doi: 10.1091/mbc.E03-04-0237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mans BJ, Anantharaman V, Aravind L, Koonin EV. Comparative genomics, evolution and origins of the nuclear envelope and nuclear pore complex. Cell Cycle. 2004;3:1612–37. doi: 10.4161/cc.3.12.1316. [DOI] [PubMed] [Google Scholar]
- 6.Osmani AH, Davies J, Liu HL, Nile A, Osmani SA. Systematic deletion and mitotic localization of the nuclear pore complex proteins of Aspergillus nidulans. Mol Biol Cell. 2006;17:4946–61. doi: 10.1091/mbc.E06-07-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 8.Iwamoto M, Mori C, Kojidani T, Bunai F, Hori T, Fukagawa T, Hiraoka Y, Haraguchi T. Two distinct repeat sequences of Nup98 nucleoporins characterize dual nuclei in the binucleated ciliate tetrahymena. Curr Biol. 2009;19:843–7. doi: 10.1016/j.cub.2009.03.055. [DOI] [PubMed] [Google Scholar]
- 9.DeGrasse JA, DuBois KN, Devos D, Siegel TN, Sali A, Field MC, Rout MP, Chait BT. Evidence for a shared nuclear pore complex architecture that is conserved from the last common eukaryotic ancestor. Mol Cell Proteomics. 2009;8:2119–30. doi: 10.1074/mcp.M900038-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tamura K, Fukao Y, Iwamoto M, Haraguchi T, Hara-Nishimura I. Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell. 2010;22:4084–97. doi: 10.1105/tpc.110.079947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neumann N, Lundin D, Poole AM. Comparative genomic evidence for a complete nuclear pore complex in the last eukaryotic common ancestor. PLoS One. 2010;5:e13241. doi: 10.1371/journal.pone.0013241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walther TC, Alves A, Pickersgill H, Loïodice I, Hetzer M, Galy V, Hülsmann BB, Köcher T, Wilm M, Allen T, et al. The conserved Nup107-160 complex is critical for nuclear pore complex assembly. Cell. 2003;113:195–206. doi: 10.1016/S0092-8674(03)00235-6. [DOI] [PubMed] [Google Scholar]
- 13.Harel A, Orjalo AV, Vincent T, Lachish-Zalait A, Vasu S, Shah S, Zimmerman E, Elbaum M, Forbes DJ. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11:853–64. doi: 10.1016/S1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 14.Loïodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–44. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rasala BA, Orjalo AV, Shen Z, Briggs S, Forbes DJ. ELYS is a dual nucleoporin/kinetochore protein required for nuclear pore assembly and proper cell division. Proc Natl Acad Sci U S A. 2006;103:17801–6. doi: 10.1073/pnas.0608484103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lutzmann M, Kunze R, Buerer A, Aebi U, Hurt E. Modular self-assembly of a Y-shaped multiprotein complex from seven nucleoporins. EMBO J. 2002;21:387–97. doi: 10.1093/emboj/21.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kampmann M, Blobel G. Three-dimensional structure and flexibility of a membrane-coating module of the nuclear pore complex. Nat Struct Mol Biol. 2009;16:782–8. doi: 10.1038/nsmb.1618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Boehmer T, Enninga J, Dales S, Blobel G, Zhong H. Depletion of a single nucleoporin, Nup107, prevents the assembly of a subset of nucleoporins into the nuclear pore complex. Proc Natl Acad Sci U S A. 2003;100:981–5. doi: 10.1073/pnas.252749899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Baï SW, Rouquette J, Umeda M, Faigle W, Loew D, Sazer S, Doye V. The fission yeast Nup107-120 complex functionally interacts with the small GTPase Ran/Spi1 and is required for mRNA export, nuclear pore distribution, and proper cell division. Mol Cell Biol. 2004;24:6379–92. doi: 10.1128/MCB.24.14.6379-6392.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu HL, De Souza CP, Osmani AH, Osmani SA. The three fungal transmembrane nuclear pore complex proteins of Aspergillus nidulans are dispensable in the presence of an intact An-Nup84-120 complex. Mol Biol Cell. 2009;20:616–30. doi: 10.1091/mbc.E08-06-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–18. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zuccolo M, Alves A, Galy V, Bolhy S, Formstecher E, Racine V, Sibarita JB, Fukagawa T, Shiekhattar R, Yen T, et al. The human Nup107-160 nuclear pore subcomplex contributes to proper kinetochore functions. EMBO J. 2007;26:1853–64. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nat Cell Biol. 2010;12:164–9. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brown JA, Bharathi A, Ghosh A, Whalen W, Fitzgerald E, Dhar R. A mutation in the Schizosaccharomyces pombe rae1 gene causes defects in poly(A)+ RNA export and in the cytoskeleton. J Biol Chem. 1995;270:7411–9. doi: 10.1074/jbc.270.13.7411. [DOI] [PubMed] [Google Scholar]
- 25.Yoon JH, Whalen WA, Bharathi A, Shen R, Dhar R. Npp106p, a Schizosaccharomyces pombe nucleoporin similar to Saccharomyces cerevisiae Nic96p, functionally interacts with Rae1p in mRNA export. Mol Cell Biol. 1997;17:7047–60. doi: 10.1128/mcb.17.12.7047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.West RR, Vaisberg EV, Ding R, Nurse P, McIntosh JR. cut11(+): A gene required for cell cycle-dependent spindle pole body anchoring in the nuclear envelope and bipolar spindle formation in Schizosaccharomyces pombe. Mol Biol Cell. 1998;9:2839–55. doi: 10.1091/mbc.9.10.2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Balasundaram D, Benedik MJ, Morphew M, Dang VD, Levin HL. Nup124p is a nuclear pore factor of Schizosaccharomyces pombe that is important for nuclear import and activity of retrotransposon Tf1. Mol Cell Biol. 1999;19:5768–84. doi: 10.1128/mcb.19.8.5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Whalen WA, Yoon JH, Shen R, Dhar R. Regulation of mRNA export by nutritional status in fission yeast. Genetics. 1999;152:827–38. doi: 10.1093/genetics/152.3.827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen XQ, Du X, Liu J, Balasubramanian MK, Balasundaram D. Identification of genes encoding putative nucleoporins and transport factors in the fission yeast Schizosaccharomyces pombe: a deletion analysis. Yeast. 2004;21:495–509. doi: 10.1002/yea.1115. [DOI] [PubMed] [Google Scholar]
- 30.Cho HJ, Hwang DK, Jung SI, Yoon JH. Schizosaccharomyces pombe nup97, which Genetically Interacts with mex67, is essential for growth and involved in mRNA export. J Microbiol. 2007;45:344–9. [PubMed] [Google Scholar]
- 31.Bae JA, Moon D, Yoon JH. Nup211, the fission yeast homolog of Mlp1/Tpr, is involved in mRNA export. J Microbiol. 2009;47:337–43. doi: 10.1007/s12275-009-0125-7. [DOI] [PubMed] [Google Scholar]
- 32.Asakawa H, Kojidani T, Mori C, Osakada H, Sato M, Ding DQ, Hiraoka Y, Haraguchi T. Virtual breakdown of the nuclear envelope in fission yeast meiosis. Curr Biol. 2010;20:1919–25. doi: 10.1016/j.cub.2010.09.070. [DOI] [PubMed] [Google Scholar]
- 33.Bilokapic S, Schwartz TU. Molecular basis for Nup37 and ELY5/ELYS recruitment to the nuclear pore complex. Proc Natl Acad Sci U S A. 2012;109:15241–6. doi: 10.1073/pnas.1205151109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu X, Mitchell JM, Wozniak RW, Blobel G, Fan J. Structural evolution of the membrane-coating module of the nuclear pore complex. Proc Natl Acad Sci U S A. 2012;109:16498–503. doi: 10.1073/pnas.1214557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tange Y, Hirata A, Niwa O. An evolutionarily conserved fission yeast protein, Ned1, implicated in normal nuclear morphology and chromosome stability, interacts with Dis3, Pim1/RCC1 and an essential nucleoporin. J Cell Sci. 2002;115:4375–85. doi: 10.1242/jcs.00135. [DOI] [PubMed] [Google Scholar]
- 36.Siniossoglou S, Wimmer C, Rieger M, Doye V, Tekotte H, Weise C, Emig S, Segref A, Hurt EC. A novel complex of nucleoporins, which includes Sec13p and a Sec13p homolog, is essential for normal nuclear pores. Cell. 1996;84:265–75. doi: 10.1016/S0092-8674(00)80981-2. [DOI] [PubMed] [Google Scholar]
- 37.Enninga J, Levay A, Fontoura BM. Sec13 shuttles between the nucleus and the cytoplasm and stably interacts with Nup96 at the nuclear pore complex. Mol Cell Biol. 2003;23:7271–84. doi: 10.1128/MCB.23.20.7271-7284.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Poloni D, Simanis V. A DMSO-sensitive conditional mutant of the fission yeast orthologue of the Saccharomyces cerevisiae SEC13 gene is defective in septation. FEBS Lett. 2002;511:85–9. doi: 10.1016/S0014-5793(01)03285-9. [DOI] [PubMed] [Google Scholar]
- 39.Strahm Y, Fahrenkrog B, Zenklusen D, Rychner E, Kantor J, Rosbach M, Stutz F. The RNA export factor Gle1p is located on the cytoplasmic fibrils of the NPC and physically interacts with the FG-nucleoporin Rip1p, the DEAD-box protein Rat8p/Dbp5p and a new protein Ymr 255p. EMBO J. 1999;18:5761–77. doi: 10.1093/emboj/18.20.5761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chadrin A, Hess B, San Roman M, Gatti X, Lombard B, Loew D, Barral Y, Palancade B, Doye V. Pom33, a novel transmembrane nucleoporin required for proper nuclear pore complex distribution. J Cell Biol. 2010;189:795–811. doi: 10.1083/jcb.200910043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang D, Vjestica A, Oliferenko S. The cortical ER network limits the permissive zone for actomyosin ring assembly. Curr Biol. 2010;20:1029–34. doi: 10.1016/j.cub.2010.04.017. [DOI] [PubMed] [Google Scholar]
- 42.Ding DQ, Tomita Y, Yamamoto A, Chikashige Y, Haraguchi T, Hiraoka Y. Large-scale screening of intracellular protein localization in living fission yeast cells by the use of a GFP-fusion genomic DNA library. Genes Cells. 2000;5:169–90. doi: 10.1046/j.1365-2443.2000.00317.x. [DOI] [PubMed] [Google Scholar]
- 43.Hayashi A, Ding DQ, Tsutsumi C, Chikashige Y, Masuda H, Haraguchi T, Hiraoka Y. Localization of gene products using a chromosomally tagged GFP-fusion library in the fission yeast Schizosaccharomyces pombe. Genes Cells. 2009;14:217–25. doi: 10.1111/j.1365-2443.2008.01264.x. [DOI] [PubMed] [Google Scholar]
- 44.Matsuyama A, Arai R, Yashiroda Y, Shirai A, Kamata A, Sekido S, Kobayashi Y, Hashimoto A, Hamamoto M, Hiraoka Y, et al. ORFeome cloning and global analysis of protein localization in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2006;24:841–7. doi: 10.1038/nbt1222. [DOI] [PubMed] [Google Scholar]
- 45.Iwamoto M, Asakawa H, Ohtsuki C, Osakada H, Koujin T, Hiraoka Y, Haraguchi T. Monoclonal antibodies recognize gly-leu-phe-gly repeat of nucleoporin nup98 of tetrahymena, yeasts, and humans. Monoclon Antib Immunodiagn Immunother. 2013;32:81–90. doi: 10.1089/mab.2012.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokunaga M, Imamoto N, Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat Methods. 2008;5:159–61. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 47.Yang Q, Rout MP, Akey CW. Three-dimensional architecture of the isolated yeast nuclear pore complex: functional and evolutionary implications. Mol Cell. 1998;1:223–34. doi: 10.1016/S1097-2765(00)80023-4. [DOI] [PubMed] [Google Scholar]
- 48.Pardo M, Nurse P. The nuclear rim protein Amo1 is required for proper microtubule cytoskeleton organisation in fission yeast. J Cell Sci. 2005;118:1705–14. doi: 10.1242/jcs.02305. [DOI] [PubMed] [Google Scholar]
- 49.Kim DU, Hayles J, Kim D, Wood V, Park HO, Won M, Yoo HS, Duhig T, Nam M, Palmer G, et al. Analysis of a genome-wide set of gene deletions in the fission yeast Schizosaccharomyces pombe. Nat Biotechnol. 2010;28:617–23. doi: 10.1038/nbt.1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Krawchuk MD, Wahls WP. High-efficiency gene targeting in Schizosaccharomyces pombe using a modular, PCR-based approach with long tracts of flanking homology. Yeast. 1999;15:1419–27. doi: 10.1002/(SICI)1097-0061(19990930)15:13<1419::AID-YEA466>3.0.CO;2-Q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Iwamoto M, Asakawa H, Hiraoka Y, Haraguchi T. Nucleoporin Nup98: a gatekeeper in the eukaryotic kingdoms. Genes Cells. 2010;15:661–9. doi: 10.1111/j.1365-2443.2010.01415.x. [DOI] [PubMed] [Google Scholar]
- 52.Davuluri G, Gong W, Yusuff S, Lorent K, Muthumani M, Dolan AC, Pack M. Mutation of the zebrafish nucleoporin elys sensitizes tissue progenitors to replication stress. PLoS Genet. 2008;4:e1000240. doi: 10.1371/journal.pgen.1000240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gao N, Davuluri G, Gong W, Seiler C, Lorent K, Furth EE, Kaestner KH, Pack M. The nuclear pore complex protein Elys is required for genome stability in mouse intestinal epithelial progenitor cells. Gastroenterology. 2011;140:1547–55, e10. doi: 10.1053/j.gastro.2011.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capelson M, Hetzer MW. The role of nuclear pores in gene regulation, development and disease. EMBO Rep. 2009;10:697–705. doi: 10.1038/embor.2009.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bukata L, Parker SL, D’Angelo MA. Nuclear pore complexes in the maintenance of genome integrity. Curr Opin Cell Biol. 2013;25:378–86. doi: 10.1016/j.ceb.2013.03.002. [DOI] [PubMed] [Google Scholar]
- 56.Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–30. doi: 10.1128/MCB.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van Werven FJ, Amon A. Regulation of entry into gametogenesis. Philos Trans R Soc Lond B Biol Sci. 2011;366:3521–31. doi: 10.1098/rstb.2011.0081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell. 2008;14:831–42. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Jong-Curtain TA, Parslow AC, Trotter AJ, Hall NE, Verkade H, Tabone T, Christie EL, Crowhurst MO, Layton JE, Shepherd IT, et al. Abnormal nuclear pore formation triggers apoptosis in the intestinal epithelium of elys-deficient zebrafish. Gastroenterology. 2009;136:902–11. doi: 10.1053/j.gastro.2008.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Iida T, Lilly MA. missing oocyte encodes a highly conserved nuclear protein required for the maintenance of the meiotic cycle and oocyte identity in Drosophila. Development. 2004;131:1029–39. doi: 10.1242/dev.01001. [DOI] [PubMed] [Google Scholar]
- 61.Senger S, Csokmay J, Akbar T, Jones TI, Sengupta P, Lilly MA. The nucleoporin Seh1 forms a complex with Mio and serves an essential tissue-specific function in Drosophila oogenesis. Development. 2011;138:2133–42. doi: 10.1242/dev.057372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Olsson M, Ekblom M, Fecker L, Kurkinen M, Ekblom P. cDNA cloning and embryonic expression of mouse nuclear pore membrane glycoprotein 210 mRNA. Kidney Int. 1999;56:827–38. doi: 10.1046/j.1523-1755.1999.00618.x. [DOI] [PubMed] [Google Scholar]
- 63.Olsson M, Schéele S, Ekblom P. Limited expression of nuclear pore membrane glycoprotein 210 in cell lines and tissues suggests cell-type specific nuclear pores in metazoans. Exp Cell Res. 2004;292:359–70. doi: 10.1016/j.yexcr.2003.09.014. [DOI] [PubMed] [Google Scholar]
- 64.D’Angelo MA, Raices M, Panowski SH, Hetzer MW. Age-dependent deterioration of nuclear pore complexes causes a loss of nuclear integrity in postmitotic cells. Cell. 2009;136:284–95. doi: 10.1016/j.cell.2008.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Meinke D, Muralla R, Sweeney C, Dickerman A. Identifying essential genes in Arabidopsis thaliana. Trends Plant Sci. 2008;13:483–91. doi: 10.1016/j.tplants.2008.06.003. [DOI] [PubMed] [Google Scholar]
- 66.Ori A, Banterle N, Iskar M, Andrés-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-L. [DOI] [PubMed] [Google Scholar]
- 68.Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–65. doi: 10.1002/(SICI)1097-0061(19960315)12:3<259::AID-YEA901>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 69.Funaya C, Samarasinghe S, Pruggnaller S, Ohta M, Connolly Y, Müller J, Murakami H, Grallert A, Yamamoto M, Smith D, et al. Transient structure associated with the spindle pole body directs meiotic microtubule reorganization in S. pombe. Curr Biol. 2012;22:562–74. doi: 10.1016/j.cub.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Asakawa H, Hiraoka Y. Live-cell fluorescence imaging of meiotic chromosome dynamics in Schizosaccharomyces pombe. Methods Mol Biol. 2009;558:53–64. doi: 10.1007/978-1-60761-103-5_4. [DOI] [PubMed] [Google Scholar]
- 71.Agard DA, Hiraoka Y, Shaw P, Sedat JW. Fluorescence microscopy in three dimensions. Methods Cell Biol. 1989;30:353–77. doi: 10.1016/S0091-679X(08)60986-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.