Fig. 1.
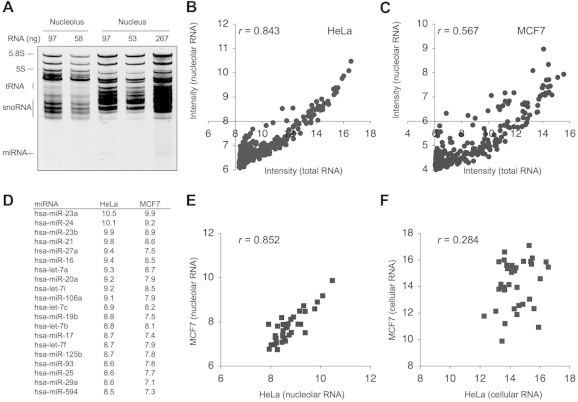
MiRNA expression profiling of nucleolar RNA. (A) Small RNA isolation and analysis by 16% denaturing PAGE. Amount of RNA loaded is indicated at the top. (B, C) MiRNA expression analysis using NCode Multi-Species array. (B) HeLa (N = 41 miRNA); (C) MCF7 (N = 48 miRNA). Scatter plot of nucleolar and total RNA intensities are shown. The nucleolar intensities were adjusted according to snoRNA expression in the respective total cellular and nucleolar fraction. Scatter plots are thresholded at 8 (HeLa) and 6 (MCF7) to exlude low intensity miRNAs. (D) Top-ranking nucleolar miRNAs in HeLa and MCF7 cells. (E, F) Scatter plots of HeLa and MCF7 nucleolar (E) and cellular (F) miRNA intensities (N = 36 miRNA). Pearson correlation coefficiencies (r) are indicated in each plot.
