Figure 3.
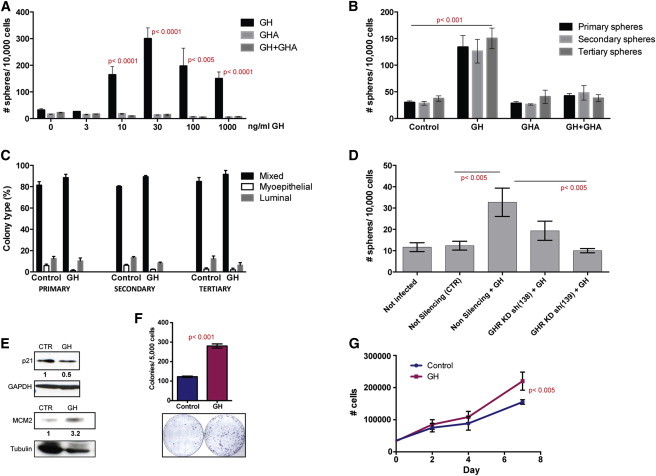
GH/GHR Signaling Is Active in Mammary Progenitor Cells
(A) A dose-dependent increase in mammosphere formation was observed upon treatment of primary HMECs with human recombinant GH. GHA abolished this effect, but did not have an effect when used alone (five independent experiments in different patient samples, each performed in triplicate, n = 5).
(B) Increased sphere formation was maintained in subsequent passages, although treatment (GH 10 ng/ml, GHA 100 ng/ml, GH+GHA) was applied only in primary culture (representative example from three independent experiments on different patient samples, each performed in triplicate, n = 3).
(C) GH treatment did not change the differentiation potential of cells composing the mammospheres. Primary, secondary, and tertiary mammospheres were dissociated, and single cells suspensions were plated at clonogenic densities on a collagen substratum in the presence of serum. After 12 days, colonies were immunostained with lineage markers (EpCAM and CD10 or CK18 and CK14). Colonies were scored based on cell composition, and percentages are shown for GH-treated and control samples (p > 0.05 for all three passages and types of colonies) (three independent experiments on two patient samples, each performed in triplicate, n = 3).
(D) Knockdown (kd) of GHR in primary HMECs did not affect sphere formation. In GHR kd cell cultures, mammosphere formation was not changed by GH treatment (representative example from four independent experiments on three patient samples, each performed in triplicate, n = 3).
(E) WB analysis showed increased level of proliferation marker MCM2 and decreased level of quiescence marker p21 after GH treatment (10 ng/ml).
(F) GH treatment (10 ng/ml) increased more than 2-fold clonogenicity of primary HMECs in adherent culture (representative example from four independent experiments on different patient samples, each performed in triplicate, n = 3).
(G) Proliferation of primary HMECs grown in adherent conditions was increased in GH-treated cultures compared to control (representative example from three independent experiments, on different patient samples, each performed in triplicate, n = 3).
Data are presented as means ± SD.
