Figure 1.
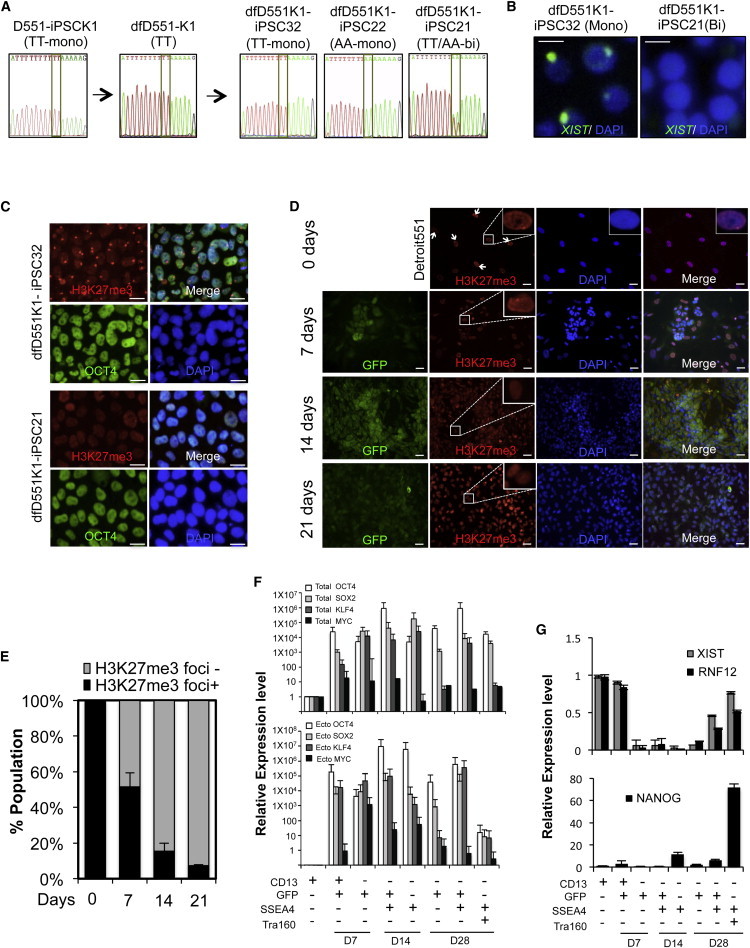
Analysis of the X Chromosome Status of Female iPSC Lines Derived from Secondary Reprogramming
(A) SNP analysis of allelic-specific expression of X-linked GRPR gene in iPSC (D551-iPSCK1) derived from Detroit 551 fibroblasts, differentiated fibroblasts (dfD551-K1), and secondary iPSC clones from dfD551-K1. When monoallelic dfD551-K1 cells having TT SNP in GRPR were reprogrammed, secondary iPSC clones that express TT SNP (dfD551K1-iPSC32), AA SNP (dfD551K1-iPSC22), or TT/AA (dfD551K1-iPSC21) were generated.
(B) RNA FISH for XIST in secondary iPSC lines at passage 12. Monoallelic dfD551-PSC32 shows XIST clouds, whereas biallelic dfD551K1-iPSC21 does not, confirming the X chromosome status. Scale bar, 20 μm.
(C) Immunostaining of H3K27me3 and OCT4 in secondary iPSC lines derived from dfD551-K1 fibroblasts. dfD551K1-iPSC32 shows H3K27me3 foci, representing the presence of inactive X chromosome, whereas dfD551K1-iPSC21 does not. Nuclei were counterstained with DAPI. Scale bar, 20 μm.
(D) Immunostaining of H3K27me3 (red) in Detroit 551 fibroblasts undergoing reprogramming on the indicated days. GFP (green) represents the expression of retrovirus-mediated reprogramming factors, and nuclei were counterstained with DAPI (blue). GFP+ cells under reprogramming lose H3K27me3 foci. Scale bar, 20 μm.
(E) Quantification of the H3K27me3 foci+ and H3K27me3 foci− cells in (A). Error bars represent mean ± SEM of three independent experiments.
(F) Relative expression of total (upper panel) and ectopic (lower panel) OCT4, SOX2, KLF4, and MYC.
(G) Relative mRNA expression of genes involved in XCI and pluripotency during reprogramming. Homogeneous populations of Detroit 551 fibroblasts undergoing reprogramming were sorted according to the expression of fibroblast marker CD13, pluripotency markers SSEA4 and TRA160, and retroviral GFP. Error bars represent mean ± SEM of three independent experiments.
