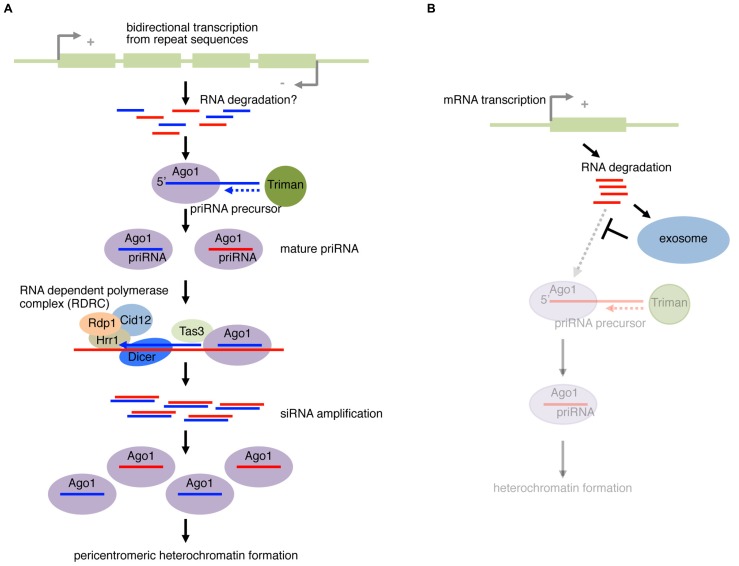
FIGURE 3.
Fission yeast priRNA pathway. (A) Bidirectional transcription from pericentromeric repeat sequences produces products that are processed into fragments, possibly through RNA degradation or other unknown mechanism. priRNA (primal RNA) precursor are bound by Ago1 and trimmed on the 3′ end by an exonuclease Triman. The loaded Ago1 with priRNA can find the nascent transcription by RNA Polymerase II in the chromatin, and recruits RNA-dependent polymerase complex (RDRC) containing Rdp1, Hrr1, and Cid12. Dicer cleaves the dsRNA to form siRNA. The siRNA can be loaded to Ago1 that continues the positive feedback cycle. The mature Ago1–siRNA complex recruits factors required for nucleation and spreading of heterochromatin to form pericentromeric heterochromatin. (B) priRNAs from mRNAs. In wild-type yeast, the exosome degrades RNA fragments. However, in mutants of exosome components, the priRNA pathway can operate more actively on mRNAs due to the lack of competition with the exosome.