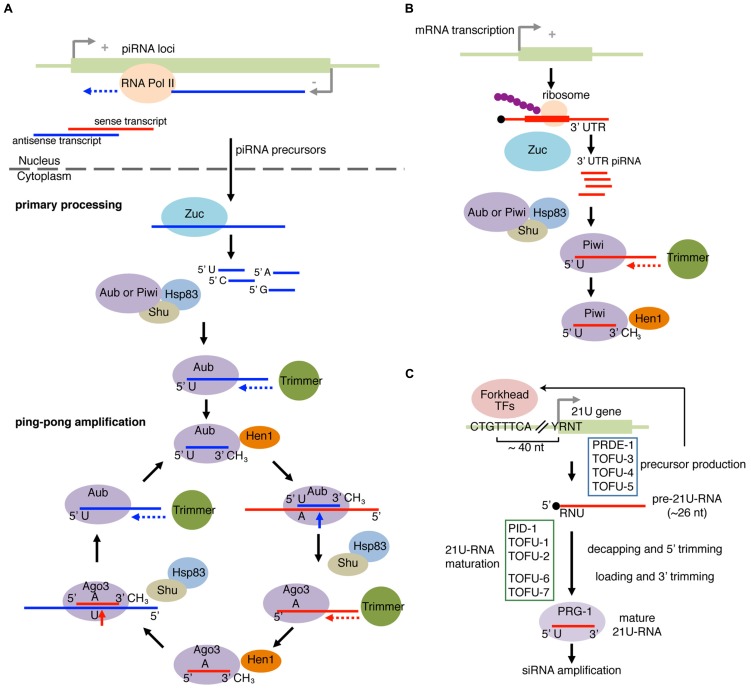
FIGURE 4.
piRNA pathway and 21U-RNA pathway. (A) Primary piRNA processing and ping-pong amplification cycle in fly. piRNA precursor transcripts are produced from piRNA loci and are cleaved by an endonuclease Zucchini (Zuc). The PIWI proteins Aubergine (Aub) or PIWI with co-chaperones select the piRNA precursor fragments with enrichment for those with 5′U. The loaded piRNA precursor fragment in PIWI or Aub is then trimmed on the 3′ end by a hypothetical exonuclease Trimmer, and 2′-O-methylated at the 2′ position of the 3′ nucleotide by the Hen1 methyltransferase. In the ping-pong amplification pathway, Aub with the mature piRNA targets transcripts bearing the complementary site. The 5′U on the mature piRNA would base pair with an A located on the complementary site. Cleavage occurs 10 nucleotides (inclusive of A) upstream of A, forming the new 5′ end. Shu and HSP83 facilitate the removal of the cleaved 5′ upstream fragment. The cleaved 3′ fragment is then passed to AGO3 and trimmed by Trimmer followed by Hen1 methylation at the 3′ end to form the secondary piRNA/AGO3 complex. This complex recognizes the complementary transcripts from the piRNA clusters and continues the cycle to amplify piRNAs. (B) piRNAs from mRNA 3′ UTR. piRNAs from mRNA 3’UTR were found in complexes containing PIWI family proteins. Zuc and Shu are required for production of 3′ UTR piRNAs, suggesting that 3′ UTR piRNAs are recognized by a mechanism similar to the primary piRNA processing pathway. (C) 21U-RNA pathway in worm produces piRNAs from the 21U gene. The majority of 21U-RNAs are generated from loci bearing the upstream CTGTTTCA and YRNT motifs (Y = pyrimidine, R = purine, N = any nucleotide). The CTGTTTCA motif is recognized by Forkhead transcription factors to enhance precursor transcription. R corresponds to the transcription start site of the 21U precursor, which is ~26 nt in length. The precursor RNA is loaded to the PIWI ortholog PRG-1. The extra nucleotides at the 5′ and 3′ ends are removed to produce mature 21U-RNAs. 21U RNA can trigger amplification of siRNA to regulate mRNAs and transposons. Recently, factors involved in 21U-RNAs were discovered. Some factors (Forkhead transcription factors, PRDE1, TOFU-3, TOFU-4, and TOFU-5) are involved in precursor biosynthesis, whereas the others (PID-1, TOFU-1, TOFU-2, TOFU-6, and TOFU-7) are involved in processing of 21U-RNA precursors.