Fig. 1.
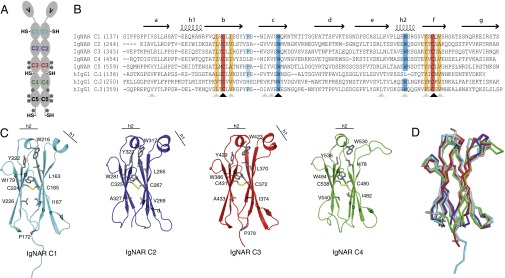
Sequence and structure of IgNAR domains C1–C4. (A) Schematic of the secreted dimeric IgNAR molecule, comprising one variable (V) and five constant (C1–C5) domains. Predicted glycosylation sites are shown as gray hexagons. Cysteines that are not part of the intradomain disulfide bridges are indicated (–SH). The secretory tail is C terminally of the C5 domain. (B) Sequence alignment of IgNAR C1–C5 with the human IgG1 HC domains CH1–CH3. Conserved cysteines are highlighted in red, and conserved hydrophobic residues of a YxCxY (Y, hydrophobic residue) motif around the disulfide bridge are highlighted in orange. Conserved tryptophans in strand c and the second helix are highlighted in blue, and the cis-proline residue in the loop between strand b and c is depicted in cyan. Secondary structure elements are indicated above the alignment. Black arrows indicate strictly conserved residues, and gray arrows homologous residues. (C) Ribbon diagram of the isolated constant IgNAR domains C1–C4 (C1, cyan; C2, blue; C3, red; C4, green; colors like in A). Residues marked in the alignment are shown in stick representation, the small helices are indicated. (D) Superposition of the IgNAR C1-4 domains (C1, cyan; C2, blue; C3, red; C4, green) on a human IgG CH3 domain (gray, Protein Data Bank ID code 1HZH).
