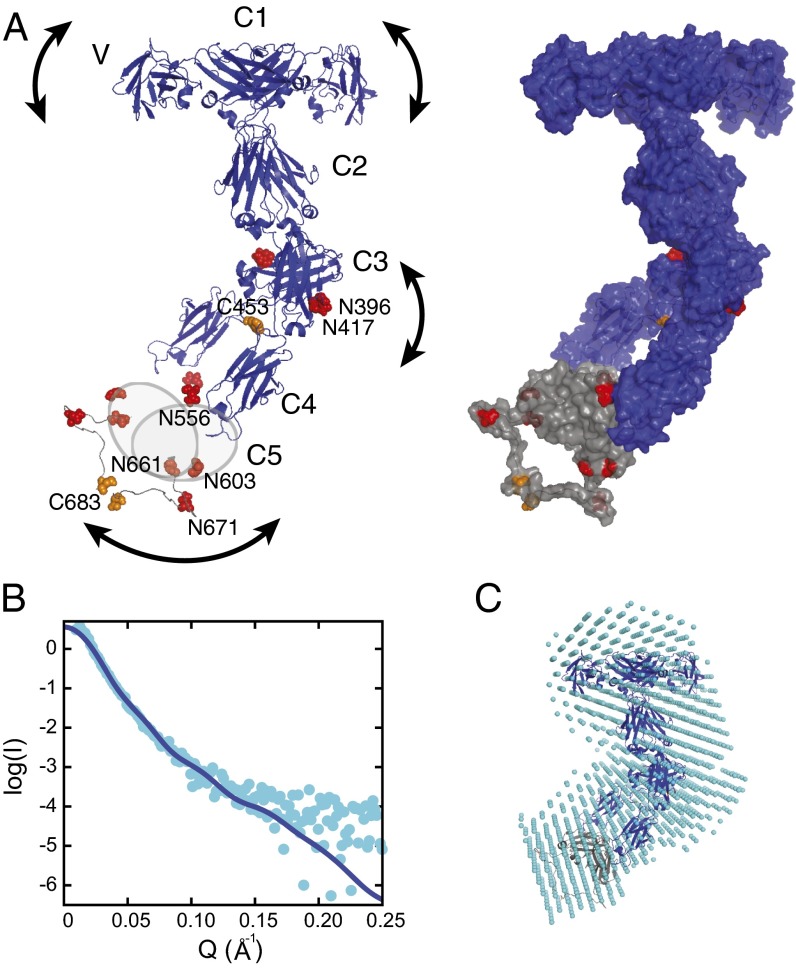
Fig. 3.
A model of the complete IgNAR antibody. (A) A structural model of full-length IgNAR based on restraints from the C1 and C3 dimer structures and intermolecular disulfide bonds, derived by docking with HADDOCK (26). (Left) Crystal structures of the variable domain (Protein Data Bank ID code 1SQ2) and constant domains C1–C4. Because of the lack of structural information for C5, it is drawn as a gray oval. Cysteines forming interchain disulfide bonds are highlighted in orange and predicted glycosylation sites in red, respectively. The arced arrows indicate flexibility within the molecule. (Right) The same model is shown in surface view, including a possible conformation of the flexible C4–C5, as adopted in the lowest energy structure of the best cluster, assessed by the HADDOCK score and SAXS χ2. (B) SAXS data of full-length IgNAR (cyan dots). The Q range above 0.25 Å−1 has been removed because of noise. The blue line corresponds to the back-calculated curve from the HADDOCK-derived structure with the best fit to the SAXS data (shown in A; χ2 = 1.43). (C) An ab initio bead model derived from the SAXS data, using DAMMIF (27), superimposed with the lowest-energy structure of the HADDOCK cluster with the best fit to the SAXS data, using SUPCOMB (49).