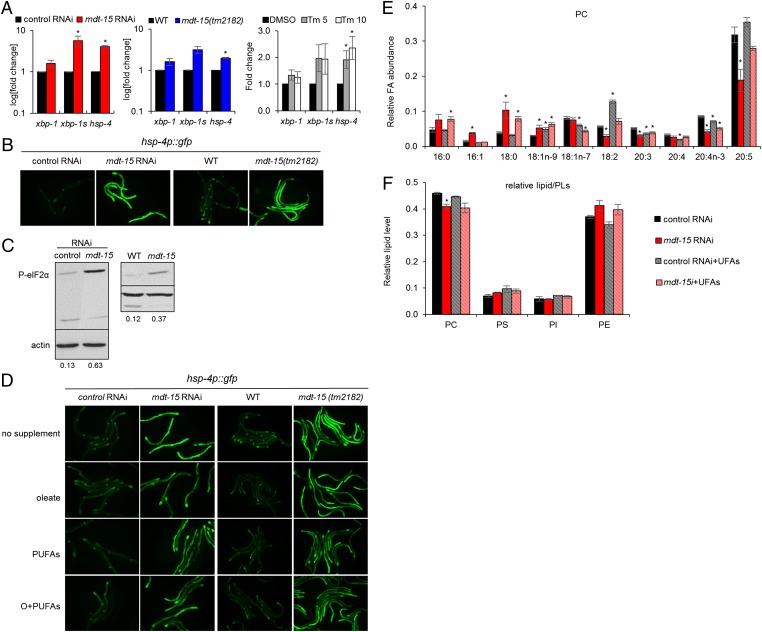
Fig. 2.
mdt-15 inactivation alters membrane lipid composition and induces the UPRER. (A) Bar graphs represent the average fold change of xbp-1, spliced xpb-1 (xbp-1s), and hsp-4 mRNA levels in control(RNAi) and mdt-15(RNAi) worms (P = 0.098, 0.049, and 0.00049, respectively) (Left), WT and mdt-15(tm2182) worms (P = 0.19, 0.063, and 0.0064, respectively) (Center), or control(RNAi) worms treated with DMSO, 5 µg/mL tunicamycin (Tm 5; P = 0.16, 0.13, 0.04, respectively) or 10 µg/mL tunicamycin (Tm 10; P = 0.31, 0.18, 0.03, respectively) (Right). n = 3 for all experiments; error bars represent SEM; *P < 0.05 (two-tailed Student t test). (B) Micrographs depict control(RNAi) and mdt-15(RNAi) worms or WT and mdt-15(tm2182) worms expressing the hsp-4p::GFP transcriptional reporter; one of four independent experiments is shown. (C) Immunoblots depict the levels of phospho-Ser51 eIF2α (P-eIF2α) and actin protein levels in control(RNAi), mdt-15(RNAi), WT, and mdt-15(tm2182) worms. The numbers represent the intensity of the P-eIF2α bands relative to corresponding actin bands. One of four independent experiments is shown. (D) Micrographs show control(RNAi) and mdt-15(RNAi) worms or WT and mdt-15(tm2182) worms expressing the hsp-4p::GFP transcriptional reporter grown without dietary supplements (no supplement), with 300 μM C18:1n-9 (oleate), with 150 μM C18:2 and 150 μM C20:5 (PUFAs), or with 150 μM C18:1n-9 and 300 μM PUFAs (O+PUFAs). (E) Bars represent the relative abundance of individual FAs in PC. (F) Bars represent the average levels of individual lipid species relative to total PLs. For E and F, lipids were extracted from L4-stage control(RNAi) or mdt-15(RNAi) worms raised in the absence or presence of dietary unsaturated FAs (UFAs; C18:1n-9, C18:2, and C20:5). n = 3; error bars represent SEM; *P < 0.05 as determined by two-tailed Student t test. For P values, see Tables S1–S3.