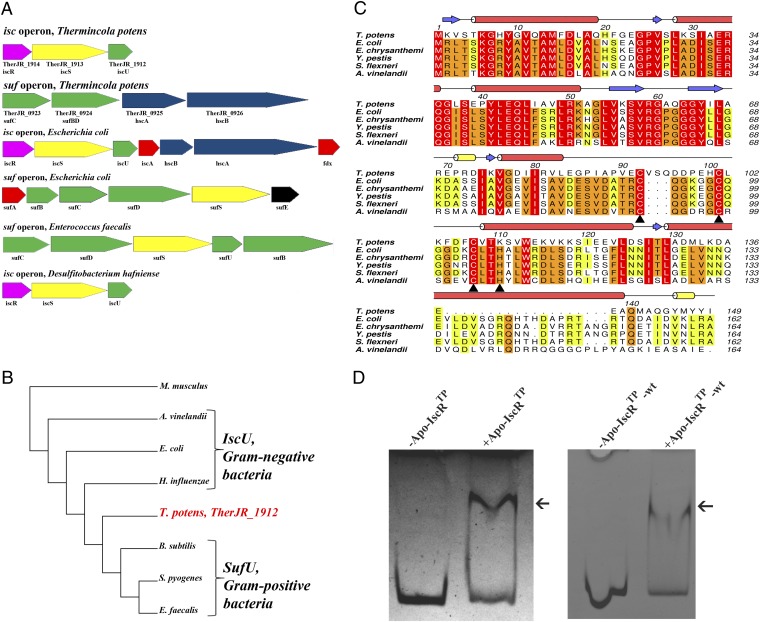
Fig. 1.
Identification of an Fe/S cluster biosynthesis regulator in T. potens. (A) T. potens possesses a unique organization of genes involved in Fe/S cluster biosynthesis, with both isc and suf operons. Colors denote gene function conservation between Gram-negative (E. coli), Gram-positive (E. faecalis), and DMRB Gram-positive bacteria (T. potens and D. hafniense). (B) The amino acid sequence of T. potens scaffold protein reveals features characteristic of the IscU-proteins from Gram-negative bacteria. Neighbor-joining phylogenetic analysis of conserved protein sequences of putative IscU-type or SufU-type proteins in both Gram-positive (T. potens, Streptococcus pyogenes, E. faecalis, B. subtilis) and Gram-negative bacteria (E. coli, Azotobacter vinelandii, Haemophilus influenza), using Mus musculus as outgroup. The sequences were aligned with three distinct alignment algorithms as implemented in ADOPS (57). The resulting cladogram places the T. potens scaffold protein between IscU proteins from Gram-negative bacteria and SufU proteins from other Gram-positive bacteria. (C) The T. potens TherJR_1914 gene codes for a protein that is highly homologous to IscR from Gram-negative bacteria. Strictly conserved amino acids are highlighted in red, and increasing residue conservation is represented by a color gradient from green to red. Alignment prepared with ClustalW (58) and colored with Aline (59). (D) T. potens apo-IscR recognizes the upstream suf operon region between genes TherJR_0922 and TherJR_0923. Incubation of either apo-IscR Cys-to-Ser mutant (apo-IscRTp) or its as-purified wild-type version (apo-IscRTp-wt) with the putative suf promoter region (suf sequence) (Table S1) resulted in a mobility-shift (arrow).