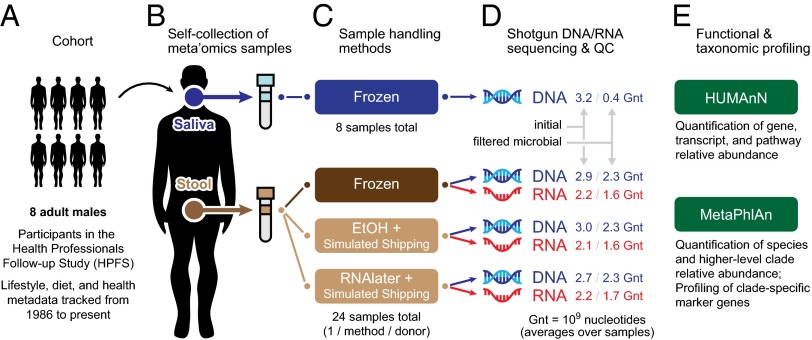
Fig. 1.
A self-sampling method compatible with metagenomic and metatranscriptomic sequencing of the human microbiome. (A) Eight participants from the HPFS cohort were recruited to assess the viability of self-collection methods in meta’omics studies and to simultaneously investigate relationships between the human oral metagenome, gut metagenome, and gut metatranscriptome. (B) Subjects self-collected samples of saliva and stool, which were returned to the laboratory. (C) Saliva samples were frozen and stool samples were tested under three conditions, including simulated shipping conditions: (i) frozen control, (ii) fixed in ethanol, and (iii) fixed in RNAlater. (D) DNA was extracted from all saliva and stool samples; RNA was extracted from stool samples only and reverse-transcribed to cDNA. All samples were then sequenced using the Illumina HiSeq platform. Raw sequence data were filtered to remove low quality and human host reads. (E) Metagenomic and metatranscriptomic read data were profiled for functional and taxonomic composition using HUMAnN (17) and MetaPhlAn (16), respectively.