Fig. 5.
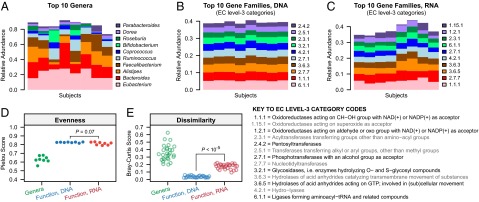
Functional diversity at the transcriptional level suggests a pattern of subject-specific metagenome regulation. (A) The 10 most abundant genera in each subject. (B) The 10 most abundant Enzyme Commission (EC) level-3 function categories in each individual, based on metagenomic measurements. (C) The 10 most abundant EC level-3 function categories in each individual, based on metatranscriptomic measurements. (D) Comparison of within-sample evenness for taxonomic and functional profiles across individuals using the Pielou metric. (E) Comparison of between-sample (β) diversity for taxonomic and functional profiles using the Bray–Curtis metric. Two-tailed P-values are based on the Wilcoxon signed-rank test. Although taxonomic profiles are highly variable among subjects, metagenomes are stable but differentially regulated at the transcriptional level.
