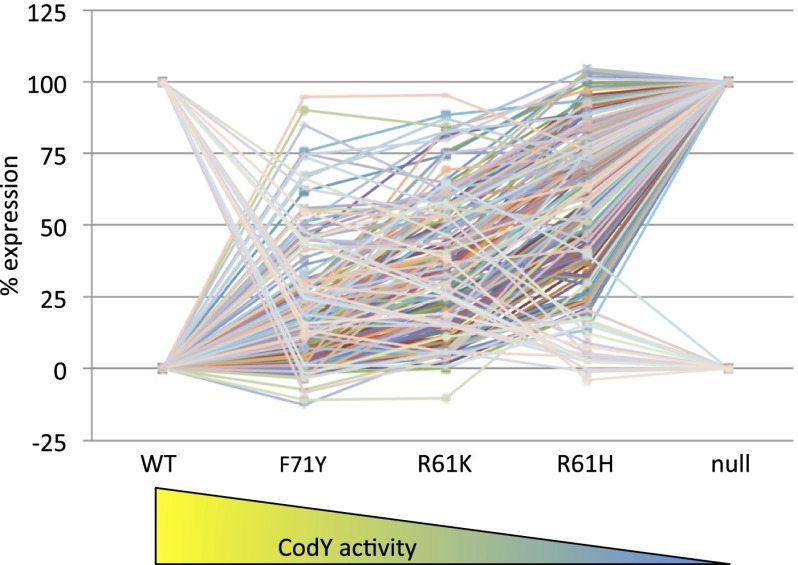
Fig. 1.
Reductions in transcription factor activity generate a diverse set of responses throughout the CodY regulon. RPKMO values for 197 differentially expressed targets with values >10 in at least one strain were plotted as a percentage of maximal expression (i.e., expression in the codY null mutant for negatively regulated genes and expression in the WT for positively regulated genes). Some CodY target genes, including adcA, frlR, rpmE2, rpsN, trpD, yczL, yrpE, ybfAB, yciABC, yjbA, and yrhP, were omitted because the data were not reproducible or the patterns were complex. Note that the positions of the point mutant strains on the x axis are arbitrary and are not meant to imply a linear relationship between position and residual CodY activity. Data points are the means of two independent experiments.