Abstract
Fluorescence-based, genetically encodable biosensors are widely used tools for real-time analysis of different biological process. Over the last few decades the number of available genetically encodable biosensors and the types of processes they can monitor has increased rapidly. In this review we aim to introduce the reader to general principles and best practices in biosensor development and highlight some of the ways in which biosensors can be used to illuminate outstanding questions of biological function. Specifically, we will focus on sensors developed for monitoring kinase activity and use them to illustrate some common considerations for biosensor design. We will describe several uses to which kinase and second-messenger biosensors have been put, and conclude with considerations for the use of biosensors once they are developed. Overall, as fluorescence-based biosensors continue to diversify and improve we expect them to continue to be widely used as reliable and fruitful tools for gaining deeper insights into cellular and organismal function.
Introduction
From the first use of fluorescently-tagged antibodies to image fixed cells through the present, scientists have wanted to “see” and thus better understand various molecular systems inside the cell. One tool that has gained traction for real-time analysis of these systems is the fluorescence-based, genetically encodable biosensor.
Early optical sensors included small molecules that bound to analytes, such as fura2 to calcium (Williams et al., 1985). Expanding the scope of biosensors to detect targets that are not easily bound by small molecules, a cAMP probe was developed in 1985 based on the dissociation of the catalytic and regulatory subunits of protein kinase A (PKA) when the kinase is activated (Adams et al., 1985). The two subunits were purified separately, tagged with the fluorophores rhodamine and fluorescein, and finally reintroduced to cells by microinjection. The dissociation of the subunits when cAMP was present caused a change in energy transfer between the two fluorophores, enabling visualization of where and when cAMP was produced for the first time. However, the probe was labor-intensive to produce and cumbersome to introduce into cells.
The advent of GFP and other fluorescent proteins solved both of these difficulties at once, and has inspired subsequent decades of optical probe design. Fluorescent proteins have proven to be a good tool for measuring in-cell activity because they are genetically encodable, bright, and available in many colors, enabling monitoring more than one target. Their genetic encodability is especially desirable in order to take advantage of the cell's own protein-synthesis machinery, introduce fluorophores with minimal disruption to the cell, and target probes to subcellular regions of interest. Furthermore, fluorescent proteins, having evolved in a cellular environment and been extensively engineered for use in mammalian cells, are compatible with cellular pHs, redox properties, and other characteristics.
After early development of genetically encoded reporters for calcium (Miyawaki et al., 1997) and cAMP (Zaccolo et al., 2000), a wide range of biosensors has been developed for monitoring different cellular events. Sensors for changes in pH and redox state; accumulation of second messengers; and activation of enzymes like kinases and phosphatases have all been reported. Kinases are a key signaling class, mediating information flow between external and internal environments or between subcellular compartments. Because of the broad reach of each activated kinase and the complex signaling cascades that lead to its activation, many kinases are key nodes in cell signaling networks.
This review aims to introduce the reader to biosensor development and application, focusing on those developed for kinases and their upstream second messengers. In the effort to understand signaling as it occurs in vivo, non-destructive technologies for monitoring signaling over time are required; biosensors are uniquely positioned to fill this need. In recent years, kinase activity reporters have been used to address a variety of systems-level questions in terminally differentiated cells and the animals from which they are derived.
In their conception and optimization, in-cell fluorescent biosensors have a great deal in common with development of small-molecule probes. Like such probes, genetically encodable biosensors are used for studies of basic biology and also to investigate the activities of many different disease-relevant signaling pathways. Such tools are effort-intensive in terms of conception, development and optimization; however, the resulting benefit of these tools especially in furthering our understanding of signaling biology outweigh the costs associated with the design process. Like any good technological product, probes are subject to quality control and are often modified based on user feedback. Thus, we introduce the “pipeline” of biosensor development and application (see figure 1). We will begin by addressing some common considerations and frequently-used blueprints for biosensor design, then survey several uses to which kinase and second-messenger biosensors have been put. We will conclude with considerations for the use of biosensors once they are developed, and the outlook of the field as a whole.
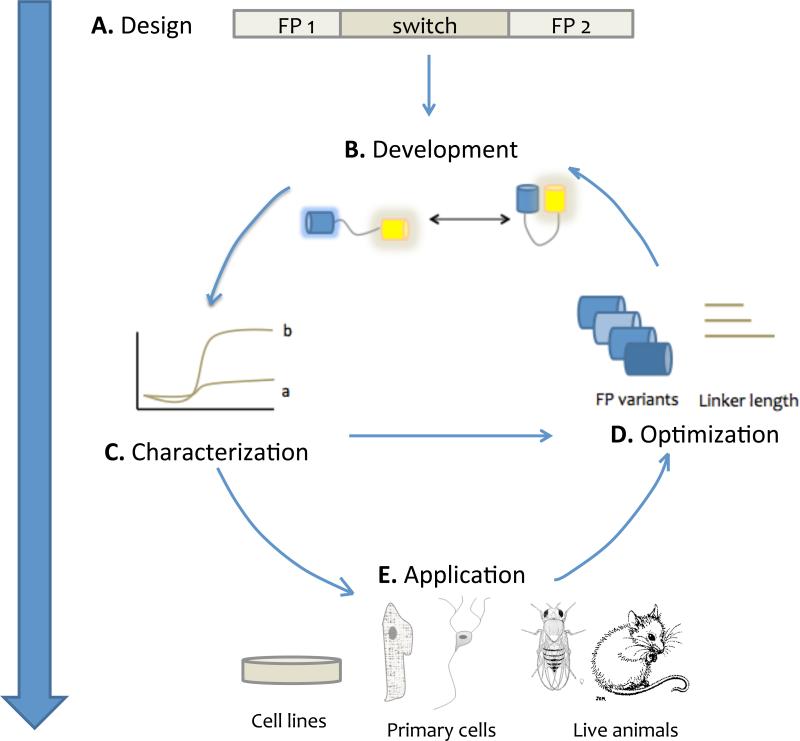
Figure 1.
The pipeline for kinase activity reporter development. (A) Design. The majority of two-fluorescent-protein reporter blueprints position the fluorescent protein pair around a molecular switch, either endogenous or engineered. (B) Development is typically an iterative process. (C ) Initial characterization of a reporter usually involves measuring its response to a known stimulus of the signal of interest; here, “a” represents an earlier version of a sensor with lower dynamic range, whereas “b” represents activity of an optimized version. (D) Optimization of a reporter may involve changing the fluorescent proteins, linker, or length/identity of the specific protein components that recognize the signal. After such optimization, new versions of a probe are compared to earlier versions. (E) Application of a probe, in cell lines (typically to investigate kinase activity and spatiotemporal regulation, or crosstalk with other pathways); primary cells (in order to explore kinase involvement in cell-type-specific behaviors); and live animals (to explore the Involvement of a kinase in specific processes or pathologies in vivo) can be both an end goal of probe development, and a spur to further optimization.
Biosensors: the pipeline
Genetically encodable probes
The general blueprint for a kinase/second messenger biosensor includes a sensing unit that senses the change of interest, and a reporting unit to indicate the sensing unit's state. To visualize a signaling event of interest, the designer of a sensor often need to identify, or engineer, a molecular switch where a change in conformation occurs in response to signaling. The reporting unit typically contains one or more fluorescent proteins. A wide variety of genetically encodable kinase activity biosensors and second messenger indicators have been developed along these lines (summarized in table 1).
Table 1.
List of genetically encoded kinase biosensors for kinase activation (by conformational change) or activity (by substrate phosphorylation).
| Target | Sensor name | References |
|---|---|---|
| Activation probes | ||
| Akt | AktAR | Gao et al., 2008, Komatsu et al. 2011 |
| GFP-PKB-RFP | Calleja et al, 2007 | |
| ReAktion | Ananthanarayan et al, 2007 | |
| B-Raf | Prin-BRaf | Terai & Matsuda, 2006 |
| C-Raf | Prin-CRaf | Terai & Matsuda, 2005 |
| Death associated protein kinase 1 (DAPK1) | Piljic et al., 2011 | |
| CamKII | Camui | Takao et al., 2005, Piljic et al., 2007, Kwok et al., 2008 |
| Erk | Miu2 | Fujioka et al., 2006 |
| P21-activated kinase 1 (PAK1) | Pakabi | Parrini et al., 2009 |
| Activity probes | ||
| Protein kinase A | AKAR | Zhang et al., 2001... Komatsu et al., 2011 |
| Abl kinase | Zhou et al., 2009 | |
| Akt | Aktus | Sasaki et al., 2003 |
| Akind | Yoshizaki et al, 2007 | |
| BKAR | Kunkel et al, 2005 | |
| AMPK | AMPKAR | Tsou et al., 2011 |
| Aurora B kinase | Aurora B sensor | Chu et al., 2011 |
| ATM kinase | Atomic | Johnson et al, 2007 |
| Protein kinase C | CKAR | Violin et al., 2003, Komatsu et al., 2011, Wu-Zhang et al., 2012 |
| Cyclin-dependent kinase 1 | Gavet & Pines, 2010 | |
| Cyclin-dependent kinase 2 | Spencer et al., 2013 | |
| Protein kinase D | DKAR | Fuchs et al., 2009, Eisler et al., 2012 |
| EGFR | Ting et al, 2001, | |
| Erk | Erkus | Sato et al, 2007 |
| EKAR | Harvey et al. 2008, Komatsu et al., 2011 | |
| REV | Xu et al., 2013 | |
| Focal adhesion kinase (FAK) | Seong et al., 2011 | |
| Inhibitor of kappa B kinase (IKK) | Olson et al., 2008 | |
| Insulin receptor | Phocus | Sato et al., 2004 |
| Sinphos | Kawai et al., 2004 | |
| c-Jun N-terminal kinase (JNK) | JNKAR | Fosbrink et al., 2010, Komatsu et al., 2011 |
| MAPK- activated protein kinase 2 (MK2) | GMB | Neininger et al., 2001 |
| Microtubule affinity regulating kinase (MARK) | Timm et al., 2010 | |
| Myosin light-chain kinase (MLCK) | exMLCK | Geguchadze et al., 2004 |
| Phosphoinositide-dependent kinase 1 (PDK1) | GFP-PDK1-RFP | Calleja et al., 2007 |
| Polo-like kinase 1 | Plk sensor | Macurek et al., 2008 |
| RSK | Eevee-RSK | Komatsu et al., 2011 |
| S6K | Eevee-S6K | Komatsu et al., 2011 |
| Stress-activated protein kinase kinase kinase | SAP3K activity reporter | Tomida et al., 2009 |
| Src | Src biosensor | Ting et al, 2001,, Wang et al, 2005, Ouyang et al., 2008 |
Sensing
One approach to generating a molecular switch is to regard the cell as a molecular toolkit—the approach used in developing the first PKA-based sensor for cAMP. The switch can be a protein or protein fragment that changes its conformation upon binding to a second messenger molecule; or it may change conformation after being enzymatically modified by the signaling enzyme of interest. Probes designed along these lines include a FRET-based sensor for the conformational change of NFAT after dephosphorylation by calcineurin (Newman et al., 2008) and a split-luciferase sensor for activity-related conformational changes in Abl kinase (Zhou et al., 2009).
If no suitable change occurs within one protein, the designer may repurpose domains from several proteins to develop an engineered switch. In the case of a kinase activity reporter, for example, a common blueprint involves the consensus substrate sequence, separated by a linker from a phospho amino acid binding domain that binds to that sequence when it is phosphorylated. The phospho amino-acid binding domain (often designated PAABD in schematic diagrams) is usually adapted from a conserved domain such as the 14,3,3 proteins; forkhead-associated (FHA) domain; two-tryptophan (WW) domain; or Src homology (SH2) domain. Sensors for activity of PKA (Zhang et al., 2001), Erk (Harvey et al., 2008), and a wide variety of other kinases use this scheme.
Switches derived from two endogenous proteins may also be left separate, in order to form a bimolecular sensor. The bimolecular sensor class is quite large and includes many sensors designed on an ad-hoc basis for studies to determine when and where two proteins of interest interact, including heteromeric binding of receptors for peptide hormones (Almabouada et al., 2013), various neurotransmitters (Borroto-Escuela et al., 2012), SNAREs (Degtyar et al., 2013) and immune signaling molecules (Hashimoto-Tane et al., 2010). In addition to providing information about the location and timing of protein interactions, bimolecular sensors can be used to determine what portions of the proteins are required for interaction (Borroto-Escuela et al., 2012) or to characterize the dissociation kinetics of the two proteins (Song et al., 2012).
Occasionally, to solve problems such as very high basal FRET, sensors initially designed in a single-polypeptide unimolecular format have been broken into bimolecular sensors by removing the linker to generate two separate proteins (Herbst et al., 2011). This modification can yield sensors with greater dynamic range, owing to better separation of FRET partners in the “off” state, but it can also introduce difficulties in data interpretation. In bimolecular versions of a probe, the donor/acceptor stoichiometry is not fixed, which can affect the signal (Jares-Erijmen and Jovin, 2003). This added variable requires the experimenter either to ascertain that the two subunits are expressed at equal levels or to use one of several correction algorithms that correct for nonhomogeneous subcellular distribution or uneven labeling before calculating FRET (Hachet-Haas et al, 2006; Deplazes et al, 2012). It has also been observed that bimolecular versions are much more sensitive to off-target binding. For example, a bimolecular version of the calcium sensor Cameleon was based on calcium induced binding between calmodulin and the calmodulin-binding domain of myosin light chain kinase (MLCK). The bimolecular version of Cameleon was suggested to have a higher tendency to bind to endogenous components (Miyawaki, 2003). Even the unimolecular version was influenced by binding of endogenous components, showing very low optical activity when endogenous calmodulin was abundant. This problem led to an effort to rationally re-engineer calmodulin and calmodulin binding peptides that bind to one another and to calcium, but clash sterically with endogenous calmodulin, based on a “bump and hole” modification of binding surfaces (Palmer et al., 2006).
If a desired switch cannot be assembled from naturally-occurring proteins or domains, it is possible to engineer a switch or part of a switch such as a binding domain. For example, in the absence of a strong and specific binding protein for phosphorylated IkBa, which is usually degraded following phosphorylation, one group used SELEX, a technique for directed evolution from a starting RNA library, to obtain a peptide that bound specifically to phospho-IkBa (Olson et al., 2008). This binding domain was tested in the context of an engineered molecular switch for an activity reporter for IKK (inhibitor of NF-κB kinase) and was found to be functional in vitro.
The conformational change of the switch can be engineered using different designs. For instance, a switch could be engineered using the design of a pseudo-ligand, which ideally binds only when the true ligand is absent. For example, an Akt-based probe for the presence of 3’ phosphoinositides used the pleckstrin homology (PH) domain from Akt, fused with a basic peptide pseudo-ligand that bound to the PH domain with low affinity. When phosphoinositides are present, the pseudo-ligand is displaced, changing the switch conformation (Ananthanarayanan et al, 2007). Based on a similar pseudoligand design, some switches are engineered to be a hybrid of genetically encoded and synthetic domains. “Snifit” is a transmembrane probe composed of a glutamate receptor with two “self-labeling” peptide tags added to the extracellular terminus. These peptides can be covalently labeled with fluorescent dyes bearing moieties that react specifically with cognate groups on the peptides (Gautier et al., 2008). In the case of Snifit, one synthetic fluorophore and another synthetic florophore conjugated with a glutamate mimic as the pseudoligand are added to label cells expressing Snifit such that FRET occurs between two fluorophores in the absence of glutamate. The pseudoligand, however, is displaced by endogenous glutamate, resulting in reduced FRET when glutamate is present in the extracellular space (Brun et al., 2012). Other synthetic strategies for kinase activity visualization have been reviewed by Rothman et al (2005).
Reporting
Once a molecular switch is selected, it must be placed within a sensor so that its binding or conformational change produces a change in optical properties. A number of design principles exist, ranging from simple intensity changes to ratiometric methods (see figure 2). Reporting domains may consist of one fluorescent protein, two peptides that combine to reconstitute a fluorescent or luminescent protein, or two fluorescent proteins.
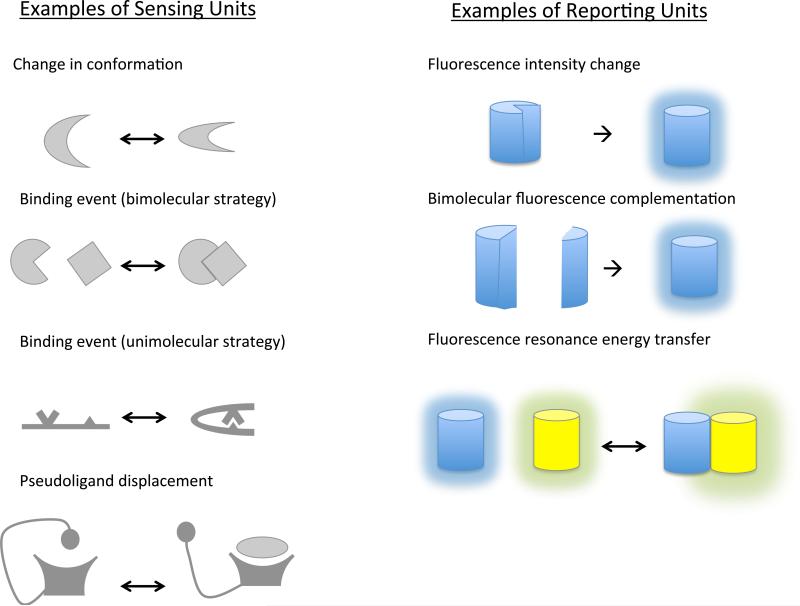
Figure 2.
Visualizing a protein's conformational change using an engineered polypeptide sequence. (A) Sensing unit: a molecular switch, usually based on endogenous sequence of a kinase or its substrate. On/off “switch” may occur endogenously, as in a conformational change or a binding event; or be engineered as in a unimolecular adaptation of a binding event or a pseudoligand displacement probe.” (B) The reporter unit generates optical readout, either by single-color fluorescence change, or by a change in resonance energy transfer.
Single fluorescent protein-based biosensors typically use a fluorescent protein with its protective beta-barrel broken, engineered so that the conformational change of the embedded molecular switch is transduced to also change the fluorescent protein's conformation and allow fluorescence to recover. This strategy has been used to create sensors for calcium, including GCAMP (Nakai et al., 2001). In one case, it has been used to indicate tyrosine phosphorylation (Kawai et al, 2004).
Bimolecular fragment complementation-based probes, feature two complementary fragments of a fluorescent protein, able to fluoresce only if they are brought into proximity, or two fragments of luciferase that reconstitute a functional luciferase when brought into proximity. These probes have been used for analysis of protein/protein interactions and kinase activity (Herbst et al., 2011); between the two, luciferase fragment complementation is more favorable for tracking reversible processes because fluorescent protein complementation is irreversible (Rebois et al., 2008).
Sensors using two fluorescent proteins typically rely on fluorescence resonance energy transfer (FRET). FRET is the non-radiative transfer of energy from one fluorophore to another via dipole-dipole interactions, resulting in reduced fluorescence from the shorter-wavelength and higher-energy “donor” fluorophore and increased emission from the longer-wavelength and lower-energy “acceptor.” Because FRET is highly distance and orientation dependent, with a sharp decrease in efficiency at nanometer-scale distances much shorter than the diffraction limit of light, it is a useful biophysical property for sensing proximity on a scale too small to be visualized as co-localization.
Flanking a molecular switch with a fluorescent protein pair competent for FRET enables monitoring of the conformation of the switch by the changes in FRET. One popular measure is FRET intensity ratio, a ratio of acceptor emission to donor emission after excitation at the donor wavelength. In addition to intensity, FRET can be measured by donor fluorescence lifetime (Padilla-Parra et al, 2012) or fluorescence anisotropy (Piston and Rizzo, 2008); these techniques, along with advantages and disadvantages of each, have been reviewed at length in the reviews listed above, as well as by Day and Davidson, 2012; Periasamy et al., 2008; and Sun et al., 2013.
Biosensor quality control and optimization
A successful probe needs to be both sensitive to and specific for the species of signaling molecule under study. Specificity for the kinase of interest needs to be verified, and the sensitivity and dynamic range of the probe generally require empirical optimization.
Once a probe is developed, and shows a satisfactory change in optical properties in response to the signal of interest, it is important to confirm that the optical change is specific to that signal. For probes based on kinase substrates, generating a quality control to rule out artifactual changes can be as easy as mutating the phosphorylated residue of the sensor to ensure that signal changes are abolished (Herbst et al., 2011). Another approach, which ensures that phosphorylation stems from the kinase of interest, is to stimulate cells expressing the kinase biosensor in the presence of an inhibitor specific to the kinase to ascertain that no spurious signal is observed (Eisler et al., 2012). The optical change may also be compared with traditional biochemical techniques like western blotting if phospho- and total protein antibodies are available for the target kinase or its substrates (Newman et al., 2008).
Many phosphorylations are carried out by a large family of related kinases with subtly different selectivity and activity; therefore, another consideration in a probe's specificity is whether it is isozyme-selective. Kajimoto et al. (2010) specialized a preexisting protein kinase C (PKC) sensor (Violin et al., 2003) based on a consensus PKC phosphorylation site, using sequences from PKCδ substrates. When the new probe was expressed alongside a panel of FP-tagged PKC isoforms, it responded specifically to PKCδ. An alternate approach to enhancing isozyme specificity is to add a selectivity-enhancing module to the sensor backbone; for example, Fosbrink et al. (2010) generated a probe selective for JNK over Erk1/2 by adding a docking domain to the shared substrate sequence. This type of optimization enables investigation of differences between closely related but differentially regulated kinases.
Once specificity has been established, a probe's sensitivity can be characterized using in vitro assays. For example, a dose-response curve showing optical changes in response to increasing analyte concentrations can be used to determine both dynamic range of the probe, in terms of response amplitude, and the concentration range of analyte that can be sensed accurately (Tantama et al., 2012). In some cases, this in-vitro step is bypassed in the development of kinase sensors because of the relative difficulty of purifying kinases in their active form; instead, the magnitude of response to a known strong stimulant of the kinase is measured. For probes with a low dynamic range or small FRET change, only very large changes in kinase or phosphatase activity can be detected. Because responses to strong stimuli in cultured cells usually far exceed responses to physiological stimuli in vivo, such probes require optimization in order to detect subtler changes. Optimization can be carried out on a semi-rational basis by tuning various factors that affect the FRET efficiency, including orientation of and distance between the two fluorophores.
Because effective energy transfer depends on alignment at an optimal orientation of the two fluorophores, minor changes during the optimization of biosensors can lead to unpredictable changes in dynamic range. One common strategy for biosensor optimization is to switch between spectrally similar fluorescent proteins in order to alter the orientation of the donor and acceptor fluorophores. For example, introduction of circularly permuted Venus, whose N and C termini are at different positions relative to the central fluorophore, can be a useful strategy to increase the dynamic range of probes (Nagai et al., 2004; Allen et al., 2006).
Distance between the fluorophores can be modulated by changes to the length of a peptide linker in a unimolecular sensor, often yielding changes in dynamic range. The linkers of unimolecular sensors have been the target of extensive optimization efforts. Several groups have worked to optimize a generic backbone using a flexible linker, which allows the fluorescent protein pair to sample many different orientations and limits signal induced changes to those dependent on distance. Although the process of adjusting linker length for optimal FRET efficiency takes time, once a suitable linker is found, it may be used for different substrate/recognition domain pairs. Komatsu et al. (2011) developed a backbone with a flexible 116 amino acid linker that improved the dynamic range for a number of kinase and GTPase sensors and was used to rapidly develop several new kinase sensors. Ibraheem et al. (2011) used a library approach to optimize linker length, comparing basal and maximally-stimulated states in replica-plated bacterial colonies. This approach allowed rapid assessment of dynamic range in a large number of variants of the probe. An alternate approach, especially useful for probes with highly structured components, is to rationally design rigid linkers to optimize fluorophore distance (Lissandron et al., 2005).
However, distance dependency of FRET may not always be easily deconvolved from orientation dependency. For example, while developing a unimolecular sensor for glutamate based on the conformational change of a glutamate-binding protein, one group observed a four-fold change in dynamic range between versions that differed in length by only one amino acid (Hires et al., 2008). They ascribed this change to the sensitivity of energy transfer to minute differences in orientation and distance, which were enhanced by the rigidity of the peptide in between the fluorescent proteins.
Publication to report on the development of a sensor is rarely the final word in its optimization (see figure 1). Even after a sensor has been introduced, updated versions are frequently released. The groups or individuals who developed a probe often continue to improve it, while users also adapt probes for their needs. In many cases, dynamic range remains relatively small, with continued optimization a desirable and ongoing process (Marvin et al, 2013; Depry et al., 2011; Kajimoto et al., 2010). Adaptations to prior probes include changing optical readouts, for example converting a probe optimized for ratiometric FRET for FLIM imaging, (Oliveira and Yasuda, 2013), and reducing sensitivity to pH and other off-target variables. Thus, optimization is most effective with specific applications in mind.
Sensor expression in primary culture and live animals
The payoff for the lengthy work of biosensor development is the ability to use the finished product to address a wide range of questions in many cell types. The fundamental question sensors uniquely answer is how various kinases function in a cellular context; although antibodies may be available to give insight into phosphorylation state, they can provide only snapshots of kinase activity. Biosensor approaches offer enhanced spatiotemporal resolution.
Often the first generation of a sensor is used to inquire into how the kinase of interest behaves in a cell line. The use of sensors to better understand such signaling has been reviewed extensively (Sipieter et al, 2013; Miyawaki, 2003; Mehta et al., 2011). Briefly put, a novel sensor can be used to explore the role of a kinase in cell activities, to study how absence or overexpression of interactors affects activity of the kinase, the location of kinase activation, and circuit analysis of interacting pathways, among other questions.
There are many cell-type-specific processes that are known to involve the activity of one or more kinases, but wherein the exact function of the kinase in question is incompletely described. In order to get that missing information in a more physiologically relevant context, primary cells and live animals are used. We will describe in detail how sensors have provided valuable information about cardiac and neuronal cell biology, then examine the emerging field of transgenic, biosensor-expressing animals.
Compartmentalized Kinase Signaling in Cardiomyocytes
Cardiac contraction is a physiological process of paramount importance, and is therefore regulated by diverse signaling pathways, many of which feature kinases in critical roles. PKA is one such kinase and is a key mediator in adrenergic signaling; stimulation of β1 or β2 adrenergic receptors activates G proteins. Gαs activates adenylyl cyclases, which produce cAMP that in turn stimulates PKA. PKA can tune cardiomyocyte contraction frequency via phosphorylation of its substrates at the sarcoplasmic reticulum, which include the SERCA regulator phospholamban and the calcium-binding adaptor protein troponin. The activation of PKA is tempered by phosphodiesterases, which limit cAMP, and some of which are activated by PKA; and A kinase anchoring proteins (AKAPs), which anchor PKA holoenzymes to specific subcellular locations.
The PKA reporter AKAR has been used, often in conjunction with cAMP reporters, to better understand adrenergic signaling in cardiomyocytes. This line of inquiry began with investigation into spatial regulation of cAMP in neonatal cardiac myocytes (Zaccolo and Pozzan, 2002), and has examined various forms of compartmentalization. For example, the differences between β1 and β2 adrenergic receptors (ARs) in stimulating PKA activity are of great interest. Tracking cAMP activity in response to extremely localized delivery of a βAR agonist with isoform-specific inhibitors, Nikolaev et al. (2010) showed that β1AR-induced cAMP signals are more diffuse whereas β2AR-induced cAMP signals are spatially confined and localized exclusively to the deep transverse tubules. In a mouse model of myocardial infarction, however, β2AR becomes distributed more diffusely over the cell surface, and produces a more diffusible cAMP wave. Related work showed that different doses of isoproterenol led to substantially different PKA dynamics as measured by AKAR (De Arcangelis et al., 2010). The increased reaction of PKA to a higher concentration of isoproterenol was traced to greater cAMP diffusibility, dependent on a dose-dependent release of PDE4 from β1ARs. Likewise using local cAMP dynamics as a clue to PDE location and activity, Sin et al. (2011) showed binding between PDE4 and a scaffolding protein that was induced by PKA phosphorylation, suggesting a negative feedback loop that keeps PKA activity within physiological range. Another check on β1AR-induced cAMP signaling is provided by AKAP79/150, which regulates β1AR recycling, and genetic deletion of which causes abnormal cAMP signaling and contraction (Li et al., 2013).
Anchored AKAR can be used to answer questions about smaller micro-environments of PKA activity; for example, a sarcoplasmic-reticulum-targeted AKAR was developed by fusing the probe to a phospholamban transmembrane sequence. This enabled the reporter to detect only the PKA activity that phospholamban and other SR membrane proteins are exposed to, rather than all PKA activity anywhere in the cell (Liu et al., 2011). This targeting innovation allowed the group to show conclusively that PKA activity is coupled to excitation and contraction. The same probe was used by Chakir et al. (2011) to investigate the response of cardiomyocytes to cardiac resynchronization therapy, a treatment for dissynchronous heart failure that was successful but whose mechanisms were poorly understood. In a model of heart failure, they showed that reduced PKA activity at the sarcomere, a slower calcium transient, and the resulting reduction in sarcomere shortening could be explained by biasing of the β2AR signaling pathway toward Gαi proteins. After resynchronization, which reversed each of these abnormalities, β2AR signaling was biased toward Gαs, restoring the ability of cAMP to accumulate after β2AR signaling, and reconnecting β2AR signaling to the sarcomere. In this system particularly, the disease state was not characterized by a change in protein levels, but rather a change in how proteins connected; a rewiring of components, rather than replacement, affected whether a signal originating from the plasma membrane could reach the sarcoplasmic reticulum. These circumstances made spatiotemporal probing especially useful.
Biosensors for other kinases have also been used to explore these kinases’ role in cardiomyocytes. For example, CaMKII is a calcium-dependent kinase regulated both by calmodulin binding and by autophosphorylation. CamKII was known to be activated downstream of β-adrenergic receptors, but its relationship to adrenergic-induced PKA activity was once murky (Grimm and Brown, 2009). The FRET sensor Camui, based on the sequence of CamKII, monitors opening of the “hinge” of the kinase in response to both of these alterations (Takao et al., 2005). Erickson et al. (2011) used this sensor to show that CamKII, like PKA, is more active in paced than quiescent cardiomyocytes; versions of the probe with mutated autophosphorylation and calmodulin-binding sites were used to dissect the mechanism of CamKII response to several neurohormonal stimuli. The probe was also shown to localize to t-tubules, indicating that CamKII likely undergoes spatial regulation similar to that of PKA.
Optical probes have contributed greatly to our understanding of both second messengers and specific kinases in β-adrenergic signaling in cardiomyocytes, and simultaneously to general rules for kinase activity, such as kinase tethering, scaffolds and compensatory pathways. However, much remains to be learned; in particular, in this oscillatory system, it would be useful to be able to visualize how phosphatase activity reverses kinase activity and resets the cell to resting state.
Compartmentalized Signaling in Neurons
Neurons are another complex primary cell type in which biosensors have enhanced our understanding of cell biology and signaling. The field of neuroscience is in the midst of an imaging revolution, harnessing the power of optics to visualize with high resolution dynamic processes in living organisms and to characterize morphological and functional responses to exquisitely targeted optogenetic stimuli (Kwon and Sabatini, 2011).
Differentiated neurons have extraordinarily complicated signaling architectures, with many levels of compartmentalization, from large divisions such as dendrite/axon/soma, to macrocompartments such as synaptic spines, to microcompartments such as those found in the neighborhood of receptors at the synapse. Each of these signaling environments maintains a unique identity and contributes to the cell's capacity for input, summing, coincidence detection, and many other computations without which cognition would be impossible. _On a cellular level, biosensor probes are useful in understanding how these computations occur, which kinases participate in spatially-restricted signaling, and what functional sub-compartments are involved in mediating inputs.
Neuron macrocompartments- dendrites and spines
On a macro scale, neurons are compartmentalized into axon, soma and dendrites, in a polarization event that occurs spontaneously in vitro. Real-time kinase and second messenger indicators have been crucial in understanding how one immature neurite is selected to become an axon while the rest mature into dendrites. An elegant study combining imaging and pharmacological manipulations of differentiating neurons showed that axon formation is dependent on a rise in cAMP and PKA activity, while dendrite differentiation is accompanied by a rise in cGMP; it was further shown that both cAMP from the developing axon and cGMP from the dendrites could feed back to inhibit cAMP production in the rest of the cell, giving a means for one and only one axon to form (Shelly et al., 2010). A followup study by the same group identified semaphorin3a as a positive regulator of dendrite differentiation that raises cGMP and reduces cAMP levels (Shelly et al., 2011).
Dendritic or synaptic spines, small protrusions from the dendrite at which excitatory synapses form often have substantially different signaling environments than the dendritic shaft from which they extend (Yasuda, 2012). Kinases are often regulated differentially in these different environments. For example, CamKII modulation of AMPA receptors and NMDA receptors contributes to long-term potentiation of synapses (reviewed at length by Lisman et al., 2012). Lee et al. (2009) showed using Camui that CamKII is activated only in individual spines where photolabile glutamate was uncaged, and that activation level is proportional to the growth of the spine. On the other hand, combining postsynaptic depolarization with the glutamate stimulation caused activity to spread into the dendrite, but only minimally into neighboring spines, highlighting the independent regulation of these compartments.
A number of studies using biosensors for proteins responsive to calcium indicate that spines have signaling capacity independent from the neuron as a whole. Wu et al. (2012) demonstrated that Alzheimer's related protein amyloid beta stimulates calcineurin most rapidly in spines. Using a bimolecular sensor comprising RFP-tagged calcineurin A and GFP-tagged calmodulin, supplemented with immunofluorescence FRET studies, the group further showed that amyloid-beta-induced calcineurin activity caused spine shrinkage and reduction of F-actin and GluR1 at the spine. Another calcium-sensitive enzyme, the protease calpain, with its activity monitored with a semi-synthetic probe made up of the protease's cleavage sequence flanked by small-molecule fluorophores, is also preferentially activated in spines (Zadran et al., 2010). In this case, however, BDNF and EGF induce spine-localized cytoskeletal rearrangements, encouraging spine growth.
Neuron microcompartments- AKAP signalosome
Scaffolding proteins enable formation of signaling complexes by clustering components of a pathway in one location to facilitate interaction. Such complexes organize signal molecules into signaling nanodomains, also known as signalosomes. In neurons, one of the most extensively studied microdomains have been centered around AKAPs. In neurons as in cardiomyocytes, PKA and cAMP sensors have been instrumental in understanding how AKAPs can modulate PKA activity by clustering signaling molecules into “nanodomains.”
For example, the PKA scaffold AKAP79/150 binds to a variety of ionotropic channels, coupling PKA activity compartmentalization to receptor sensitivity. Overexpression of AKAP79/150 reduces the cAMP response to calcium influx in neurons, reducing PKA activity (Willoughby et al., 2010). Among the functions of the scaffold shown using AKAR and related probes are retention of PKA near M-type receptors (Bal et al), retention of adenylyl cyclases near AMPA receptors (Efendiev et al., 2010) and retention of adenylyl cyclase near the calcium channel trpv1 (Efendiev et al., 2013). The trpv1 channel is of particular interest because AKAP79/150 binding is required for its phosphorylation by PKA and PKC. A FRET study using a bimolecular sensor for AKAP/trpv1 binding showed that calcium influx, following receptor stimulation, reduces binding in a negative feedback loop (Chaudhury et al., 2011). Meanwhile, AKAP79 also binds to calcineurin, opposing channel opening in some channel species (Oliveria et al., 2012). This counterbalance underlines again the exquisite balance of kinase and phosphatase activity in regulating signaling.
Transgenic biosensor animals
While most of the experiments described above were carried out in transiently transfected primary cell cultures, studies of PKA in neurons have recently become available in a whole-brain context in live, behaving animals. One study used a fly line inducibly expressing AKAR, which was imaged via two-photon FLIM, to understand why the mutants rutabaga and dunce have defects in aversive learning (Gervasi et al., 2010). The authors found that rutabaga, a calcium-sensitive adenylyl cyclase, is a coincidence detector for octopaminergic and dopaminergic stimuli that are both below threshold levels, facilitating associative learning; mutations in rut slowed the accumulation of PKA activity. On the other hand, mutations to dunce, a phosphodiesterase, caused a slightly higher PKA response than the wild-type; the phosphodiesterase plays a role in restriction of dopamine-responsive PKA activity to specific brain regions.
This is a good example of the potential of kinase and phosphatase studies in vivo to further our understanding of the connections between subcellular signaling circuits and behavioral output. For translational purposes, mammalian systems such as mouse are appealing targets for similar in vivo studies. However, there are a number of challenges for in vivo kinase studies, particularly in living mice. Because of the propensity of tissue to scatter light, special imaging techniques may be needed for effective kinase activity visualization in vivo. Many studies use two-photon fluorescence imaging on surgically exposed tissues. Two-photon imaging, which uses two long-wavelength beams to excite only within a focal spot, offers better tissue penetration and reduced photobleaching compared to shorter wavelength illumination.
Expression level is a constant challenge in the development of biosensor-expressing animals. Sensors can be silenced or recombined out of the genome (Yamaguchi et al., 2011), and they sometimes appear unpredictably in only a few tissues. For example, in a double-transgenic mouse line carrying two halves of a bimolecular reporter, both driven by the same ostensibly ubiquitous reporter, one was expressed reasonably brightly in a number of tissue types, whereas the second appeared only in testes (Audet et al., 2010). When expression is too low, effective FRET studies cannot be carried out because of extremely low signal-to-noise ratios. Overexpression, however, may cause embryonic lethality or perturbation of the signaling system beyond physiological relevance (Hara et al., 2004). Although transgenic calcium-sensing mice, including some that express FRET-based sensors such as Cameleon (Hara et al, 2004), have existed for some time, mice expressing kinase sensors have only recently became available.
A recent effort using cytoplasmic mRNA injection yielded mice ubiquitously expressing AKAR, EKAR and nucleus-targeted EKAR (Kamioka et al., 2012). These were used for several proof-of-principle studies, including comparative analysis of PKA and Erk responses to laser ablation of epidermal tissues. The authors propose that these mice may be useful for studying pharmacodynamics in vivo.
The same mouse lines have also proven useful, albeit in a tissue-culture explant system, for studying relationships between signaling circuits. Gut explants from AKAR and EKAR mice were used to study the relationships between pro-migration and anti-migration signaling circuits in organ-cultured enteric nervous system progenitor cells (Goto et al., 2013). By watching the relationship between location and kinase or GTPase activity in real time in a network of neural progenitor cells advancing along the gut, the authors determined that Rac1 and Cdc42 activity were highest toward the leading edge, both within and between cells, whereas PKA activity was lower in the migrating cells and higher in the stationary network cells. The authors were also able to show that some kinases, such as JNK, that were purportedly required for migration, do not seem to affect its velocity or direction.
Blending activity-based biosensing with two-photon techniques for chronic invivo neuron imaging, Mower et al. (2013) showed that CamKII, mentioned above for its role in spine signaling in dissociated cultures, also plays a key role in spine and synapse maintenance in living brains. In ferrets expressing Camui delivered by viral vector, they determined that irrespective of starting spine size, dendritic spines with low basal CamKII activity were more likely to be lost after sensory deprivation than spines with higher starting CamKII.
Outlook
As studies with just a few key kinases make clear, kinase activity reporters can be an extraordinarily useful tool for understanding the target enzyme and its adaptors, modulators and cofactors. As new probes are developed, and existing probes optimized for primary culture and in-vivo imaging, we anticipate similarly diverse findings in many pathways. Progress is underway at many points in the pipeline, from target selection through optimization to live-animal applications.
For the purposes of optimization, structural studies of extant probes will be important to advance our understanding of sensor technology and settle questions about how probes function. For example, a recent small-angle X-ray scattering (SAXS) study with the calcium sensor TN-XXL showed that distance played a greater role than orientation in the FRET efficiency of the calcium-free form of the sensor (Geiger et al., 2012). Further insight into what specific conformational changes lead to the greatest FRET change will be beneficial for future probe design; another recent study used SAXS assessment of the distance between fluorescent proteins in a probe's off-state and on-state to predict the maximal expected FRET change (Mertens et al., 2012). Such information will enable rational design and modification of future probes.
Novel strategies present an avenue for multiplex visualization of several enzymes at a time. For example, three-fluorescent-protein FRET (reviewed by Depry et al., 2013) enables simultaneous visualization of two FRET-based biosensors. Likewise, multicolor bimolecular fluorescence complementation (Waadt et al., 2008) may allow characterization of complexes that regulate kinase signaling.
Some of these genetically encoded biosensors have been used in compound screens in a 96-well format (Allen et al., 2006). Further enhancement of the dynamic range could facilitate this important application in cellular context to complement FRET-based inhibitor screens using synthetic sensors outside of the cellular context (e.g., Gratz et al., 2011). Another application of the genetically encoded sensors where we expect to see further advancement is inhibitor candidate validation (e.g., Timm et al., 2011), including validation of inhibitors’ isoform specificity (Tsalkova et al., 2012), and analysis of pharmacokinetics in in vitro or in vivo models (Nobis et al., 2013).
Recent progress in live-animal visualization of kinase activity is encouraging, but more remains to be done. The reduction in signal-to-noise ratio and dynamic range observed in many in vivo systems may necessitate probe optimization and modification, for example, red-shifting of sensors (Lohse et al., 2012). It is important to keep in mind that improvements in one parameter may come at the cost of others, and that different versions of a sensor may be best suited to different applications. We look to recent advances in live animal studies with genetically encoded calcium indicators (reviewed by Palmer et al., 2011) as a model for the way forward with kinase sensors. In particular, iterative optimization has proven successful for genetically encoded calcium indicators, where often an early version shows very weak response in live animals, but subsequent versions work much better.
Advances in in vivo imaging technology, used in combination with improved probes, will further expand the reach of kinase and phosphatase sensors. In vivo imaging that can be carried out over developmental time-courses, using lower illumination; in intact tissues, using higher-penetrance illumination techniques; and in freely behaving animals, using flexible optical devices, will be key to our understanding of signaling in the context of systems that are difficult to model in vitro. Emerging technologies for genome editing such as TALEN and CRISPR systems (Bedell et al 2012; Ran et al., 2013) should facilitate the introduction of these biosensors into various systems, further expanding the scope of their applications.
In conclusion, fluorescence-based biosensors have come a long way, and continue to shine a light on cellular activity. We anticipate a fruitful future for these tools, yielding many more lessons in kinase signaling regulation.
Acknowledgments
We gratefully acknowledge the assistance of Welch Library informationist Rob Wright, MLS, in constructing literature searches with controlled vocabulary, and that of Ambhiganath Ganesan and Fabian Hertel in commenting on drafts of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams, Harootunian, Buechler, Taylor, Tsien Fluorescence ratio imaging of cyclic AMP in single cells. Nature. 1991;349:694–7. doi: 10.1038/349694a0. [DOI] [PubMed] [Google Scholar]
- Allen, Zhang Subcellular dynamics of protein kinase A activity visualized by FRET-based reporters. Biochem Biophys Res Comm. 2006;348:716–21. doi: 10.1016/j.bbrc.2006.07.136. [DOI] [PubMed] [Google Scholar]
- Almabouada, Diaz-Ruiz, Rabanal-Ruiz, Peinado, Vazquez-Martinez, Malagon Adiponectin receptors form homomers and heteromers exhibiting distinct ligand binding and intracellular signaling properties. J Biol Chem. 2013;288:3112–25. doi: 10.1074/jbc.M112.404624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ananthanarayanan, Fosbrink, Rahdar, Zhang Live-cell molecular analysis of Akt reveals roles for activation loop phosphorylation. J Biol Chem. 2007;282:36634–41. doi: 10.1074/jbc.M706227200. [DOI] [PubMed] [Google Scholar]
- Ananthanarayanan, Ni, Zhang Signal propagation from membrane messengers to nuclear effectors revealed by reporters of phosphoinositide dynamics and Akt activity. PNAS. 2005;102:15081–6. doi: 10.1073/pnas.0502889102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Audet, Lagace, Silversides, Bouvier Protein-protein interactions monitored in cells from transgenic mice using bioluminescence resonance energy transfer. FASEB J. 2010;24:2829–38. doi: 10.1096/fj.09-144816. [DOI] [PubMed] [Google Scholar]
- Bal, Zhang, Hernandez, Zaika, Shapiro Ca2+/calmodulin disrupts AKAP79/150 interactions with KCNQ (M-Type) K+ channels. J Neurosci. 2010;30:2311–23. doi: 10.1523/JNEUROSCI.5175-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedell, Wang, Campbell, Poshusta, Starker, Krug, Tan, Penheiter, Ma, Leung, Fahrenkrug, Carlson, Voytas, Clark, Essner, Ekker In vivo genome editing using a high-efficiency TALEN system. Nature. 2012;491:114–8. doi: 10.1038/nature11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borroto-Escuela, Romero-Fernandez, Mudo, Perez-Alea, Ciruela, Tarakanov, Narvaez, Di Liberto, Agnati, Belluardo, Fuxe Fibroblast growth factor receptor 1- 5-hydroxytryptamine 1A heteroreceptor complexes and their enhancement of hippocampal plasticity. Biol Psych. 2012;71:84–91. doi: 10.1016/j.biopsych.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Brun, Tan, Griss, Kielkowska, Reymond, Johnsson A semisynthetic fluorescent sensor protein for glutamate. J Am Chem Soc. 2012;134:7676–8. doi: 10.1021/ja3002277. [DOI] [PubMed] [Google Scholar]
- Calebiro, Nikolaev, Gagliani, de Filippis, Dees, Tacchetti, Persani, Lohse Persistent cAMP-signals triggered by internalized GPCRs. PLoS Biology. 2009;7:e1000172. doi: 10.1371/journal.pbio.1000172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calleja, Alcor, Laguerre, Park, Vojnovic, Hemmings, Downward, Parker, Larijani Intramolecular and intermolecular interactions of protein kinase B define its activation in vivo. PLoS Biology. 2007;5:e95. doi: 10.1371/journal.pbio.0050095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro, Gervasi, Guiot, Cavelini, Nikolaev, Paupardin-Tritsch, Vincent Type 4 phosphodiesterase plays different integrating roles in different cellular domains in pyramidal cortical neurons. J Neuroscience. 2010;17:6143–51. doi: 10.1523/JNEUROSCI.5851-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakir, Depry, Dimaano, Zhu, Vanderheyden, Bartunek, Abraham, Tomaselli, Liu, Xiang, Zhang, Takimoto, Dulin, Xiao, Zhang, Kass Galphas-biased beta2-adrenergic receptor signaling from restoring synchronous contraction in the failing heart. Sci Transl Med. 2011;3:100. doi: 10.1126/scitranslmed.3001909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury, Bal, Belugin, Shapiro, Jeske AKAP150-mediated TRPV1 sensitization is disrupted by calcium/calmodulin. Mol Pain. 2011;7:34–8069-7-34. doi: 10.1186/1744-8069-7-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, Sabatini Signaling in dendritic spines and spine microdomains. Curr Op Neurobio. 2012;22:389–96. doi: 10.1016/j.conb.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu, Yao, Wang, Wang, Wang, Zhang, Huang, Ke, Ding, Yao Aurora B kinase activation requires surviving priming phosphorylation by PLK1. J Mol Cell Bio. 2011;3:260–7. doi: 10.1093/jmcb/mjq037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day, Davidson Fluorescent proteins for FRET microscopy: monitoring protein interactions in living cells. BioEssays. 2012;34:341–50. doi: 10.1002/bies.201100098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Arcangelis, Liu, Zhang, Soto, Xiang Equilibrium between adenylyl cyclase and phosphodiesterase patterns adrenergic agonist dose-dependent spatiotemporal cAMP/protein kinase A activities in cardiomyocytes. Mol Pharmacol. 2010;78:340–9. doi: 10.1124/mol.110.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedecker, De Schryver, Hofkens Fluorescent Proteins: Shine on, You Crazy Diamond. J Am Chem Soc. 2013;135:2387–402. doi: 10.1021/ja309768d. [DOI] [PubMed] [Google Scholar]
- Degtyar, Hafez, Bray, Zucker Dance of the SNAREs: Assembly and Rearrangements Detected with FRET at Neuronal Synapses. J Neurosci. 2013;33:5507–23. doi: 10.1523/JNEUROSCI.2337-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deplazes, Jayatilaka, Corry ExiFRET: flexible tool for understanding FRET in complex geometries. J Biomed Optics. 2012;17:011005, 1–11. doi: 10.1117/1.JBO.17.1.011005. [DOI] [PubMed] [Google Scholar]
- Depry, Allen, Zhang Visualization of PKA activity in plasma membrane microdomains. Mol Biosystems. 2011;7:52–8. doi: 10.1039/c0mb00079e. [DOI] [PubMed] [Google Scholar]
- Depry, Mehta, Zhang Multiplexed visualization of dynamic signaling networks using genetically encoded fluorescent protein-based biosensors. Pflugers Eur J Physiol. 2013;465 doi: 10.1007/s00424-012-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efendiev, Bavencoffe, Hu, Zhu, Dessauer Scaffolding by A kinase anchoring protein enhances functional coupling between adenylyl cyclase and TRPV1 channel. J Biol Chem. 2013;288:3929–37. doi: 10.1074/jbc.M112.428144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efendiev, Samelson, Nguyen, Phatarpekar, Baameur, Scott, Dessauer AKAP79 interacts with multiple adenylyl cyclase (AC) isoforms and scaffolds AC5 and -6 to alpha-amino-3-hydroxyl-5-methyl-4-isoxazole-propionate (AMPA) receptors. J Biol Chem. 2010;285:14450–8. doi: 10.1074/jbc.M110.109769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisler, Fuchs, Pfizenmaier, Hausser G-PKDrep-live, a genetically encoded FRET reporter to measure PKD activity at the trans-Golgi-network. Biotech J. 2012;7:148–54. doi: 10.1002/biot.201100273. [DOI] [PubMed] [Google Scholar]
- Erickson, Patel, Ferguson, Bossuyt, Bers Fluorescence resonance energy transfer-based sensor Camui provides new insight into mechanisms of calcium/calmodulin-dependent protein kinase II activation in intact cardiomyocytes. Circulation Research. 2011;109:729–38. doi: 10.1161/CIRCRESAHA.111.247148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa, Ramberg, Gatsinzi, Samuelsson, Zhang, Iverfeldt, Hallberg Anchored FRET sensors detect local caspase activation prior to neuronal degeneration. Mol Neurodegen. 2011;6 doi: 10.1186/1750-1326-6-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fosbrink, Aye-Han, Cheong, Levchenko, Zhang Visualization of JNK activity dynamics with a genetically encoded fluorescent biosensor. PNAS. 2010;107:5459–64. doi: 10.1073/pnas.0909671107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs, Eisler, Link, Schlicker, et al. A Golgi PKD activity reporter reveals a crucial role of PKD in nocodazole-induced Golgi dispersal. Traffic. 2009;10:858–867. doi: 10.1111/j.1600-0854.2009.00918.x. [DOI] [PubMed] [Google Scholar]
- Fujioka, Terai, Itoh, Aoki, Nakamura, Kuroda, Nishida, Matsuda Dynamics of the Ras/Erk MAPK cascade as monitored by fluorescent probes. J Biol Chem. 2006;281:8917–26. doi: 10.1074/jbc.M509344200. [DOI] [PubMed] [Google Scholar]
- Gao, Zhang Spatiotemporal analysis of differential Akt regulation in plasma membrane microdomains. Mol Bio Cell. 2009;19:4366–73. doi: 10.1091/mbc.E08-05-0449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gautier, Juillerat, Heinis, Correa, Kindermann, Beaufils, Johnsson An engineered protein tag for multiprotein labeling in living cells. Chem and Biol. 2008;15:128–36. doi: 10.1016/j.chembiol.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Gavet, Pines Activation of cyclin B1-Cdk1 synchronizes events in the nucleus and cytoplasm at mitosis. J Cell Biol. 2010;189:247–59. doi: 10.1083/jcb.200909144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geguchadze R, Zhi G, Lau KS, Isotani E, Persechini A, Kamm Ke, Stull JT. Quantitative measurements of clciu,m/calmodulin binding and activation of myosin light chain kinase in cells. FEBS letters. 2004;557:121–124. doi: 10.1016/s0014-5793(03)01456-x. [DOI] [PubMed] [Google Scholar]
- Geiger, Russo, Gensch, Thestrup, Becker, Hopfner, Griesinger, Witte, Griesbeck Correlating calcium binding, Forster resonance energy transfer, and conformational change in the biosensor TN-XXL. Biophys J. 2012;102:2401–10. doi: 10.1016/j.bpj.2012.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gervasi, Tchenio, Preat PKA dynamics in a Drosophila learning center: coincidence detection by rutabaga adenylyl cyclase and spatial regulation by dunce phosphodiesterase. Neuron. 2010;65:516–29. doi: 10.1016/j.neuron.2010.01.014. [DOI] [PubMed] [Google Scholar]
- Ghigo, Perino, Mehel, Zahradnikova, Morello, Leroy, Nikolaev, Damilano, Cimino, De Luca, Richter, Westenbroek, Catterall, Zhang, Yan, Conti, Gomez, Vandecasteele, Hirsch, Fischmeister Phosphoinositide 3-Kinase gamma Protects Against Catecholamine-Induced Ventricular Arrhythmia Through Protein Kinase A-Mediated Regulation of Distinct Phosphodiesterases. Circulation. 2012;126:2073. doi: 10.1161/CIRCULATIONAHA.112.114074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz, Gotz, Jose A FRET-based microplate assay for human protein kinase CK2, a target in neoplastic disease. J Enz Inhib and Med Chem. 2010;25:234–9. doi: 10.3109/14756360903170038. [DOI] [PubMed] [Google Scholar]
- Grimm, Brown Beta-adrenergic receptor signaling in the heart: role of CaMKII. J Mol Cell Cardiol. 2009;48:322–30. doi: 10.1016/j.yjmcc.2009.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, Sumiyama, Kamioka, Nakasyo, Ito, Iwasaki, Enomoto, Matsuda GDNF and Endothelin 3 Regulate Migration of Enteric Neural Crest-Derived Cells via Protein Kinase A and Rac1. J Neuroscience. 2013;33:4901–12. doi: 10.1523/JNEUROSCI.4828-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto, Hoshino, Matsuda, Nakamura Phosphorylation of STEF/Tiam2 by protein kinase A is critical for Rac1 activation and neurite outgrowth in dibutyryl cAMP-treated PC12D cells. Mol Biol Cell. 2011;22:1780–90. doi: 10.1091/mbc.E10-09-0783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacht-Haas, Converset, Marchal, Matthes, Gloria, Galzi, Lecat FRET and colocalization analyzer—a method to validate measurements of sensitized emission FRET acquired by confocal microscopy and available as an ImageJ plug-in. Microscopy Research and Technique. 2006;69:941–56. doi: 10.1002/jemt.20376. [DOI] [PubMed] [Google Scholar]
- Hara, Bindokas, Lopez, Kaihara, Landa, Harbeck, Roe Imaging endoplasmic reticulum calcium with a fluorescent biosensor in transgenic mice. Am J Physiol Cell Physiol. 2004;287:932–8. doi: 10.1152/ajpcell.00151.2004. [DOI] [PubMed] [Google Scholar]
- Harvey, Ehrhardt, Cellurale, Zhong, Yasuda, Davis, Svoboda A genetically encoded fluorescent sensor of ERK activity. Proc Nat Acad Science. 2008;105:19264–9. doi: 10.1073/pnas.0804598105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto-Tane, Yokosuka, Ishihara, Sakuma, Kobayashi, Saito T-cell receptor microclusters critical for T-cell activation are formed independently of lipid raft clustering. Mol Cell Biol. 2010;30:3421–9. doi: 10.1128/MCB.00160-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbst, Allen, Zhang Luminescent kinase activity biosensors based on a versatile bimolecular switch. J Am Chem Soc. 2011;133:5676–9. doi: 10.1021/ja1117396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hires, Zhu, Tsien Optical measurement of synaptic glutamate spillover and reuptake by linker optimized glutamate-sensitive fluorescent reporters. Proc Nat Acad Sci. 2008;105:4411. doi: 10.1073/pnas.0712008105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibraheem, Yap, Ding, Campbell A bacteria colony-based screen for optimal linker combinations in genetically encoded biosensors. BMC Biotech. 2011;11 doi: 10.1186/1472-6750-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares-Erijman, Jovin FRET imaging. Nature Biotech. 2003;21:1387–95. doi: 10.1038/nbt896. [DOI] [PubMed] [Google Scholar]
- Johnson, You, Hunter Monitoring ATM kinase activity in living cells. DNA Repair. 2007;6:1277–84. doi: 10.1016/j.dnarep.2007.02.025. [DOI] [PubMed] [Google Scholar]
- Kajimoto, Sawamura, Tohyama, Mori, Newton Protein kinase C {delta}-specific activity reporter reveals agonist-evoked nuclear activity controlled by Src family of kinases. J Biol Chem. 2010;285:41896–910. doi: 10.1074/jbc.M110.184028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamioka, Sumiyama, Mizuno, Sakai Y, Hirata, Kiyokawa, Matsuda Live imaging of protein kinase activities in transgenic mice expressing FRET biosensors. Cell structure and function. 2012;37:65–73. doi: 10.1247/csf.11045. [DOI] [PubMed] [Google Scholar]
- Kawai, Sato, Umezawa Single color fluorescent indicators of protein phosphorylation for multicolor imaging of intracellular signal flow dynamics. Anal Chem. 2004;76:6144–9. doi: 10.1021/ac040037s. [DOI] [PubMed] [Google Scholar]
- Komatsu, Aoki, Yamada, Yukinaga, Fujita, Kamioka, Matsuda Development of an optimized backbone of FRET biosensors for kinases and GTPases. Mol Biol Cell. 2011;22:4647. doi: 10.1091/mbc.E11-01-0072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, Toker, Tsien, Newton Calcium-dependent regulation of protein kinase D revealed by a genetically encoded kinase activity reporter. J Biol Chem. 2007;282:6733042. doi: 10.1074/jbc.M608086200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok, Lee, Sanchez, Hazlett, Gratton, Hayashi Genetically encoded probe for fluorescence lifetime imaging of CamKII activity. BBRC. 2008;369:519–25. doi: 10.1016/j.bbrc.2008.02.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon, Sabatini Glutamate induces de novo growth of functional spines in developing cortex. Nature. 2011;474:100–4. doi: 10.1038/nature09986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, Escobedo-Lozoya, Szatmari, Yasuda Activation of CAMKII in single dendritic spines during long-term potentiation. Nature. 2009;458:299. doi: 10.1038/nature07842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Nooh, Bahouth Role of AKAP79/150 in beta-1-adrenergic receptor trafficking and signaling in mammalian cells. J Biol Chem. 2013 doi: 10.1074/jbc.M113.470559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisman, Yasuda, Raghavachari Mechanisms of CamKII action in long-term potentiation. Nat Rev Neurosci. 2012;13:169–82. doi: 10.1038/nrn3192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lissandron, Terrin, Collini, D'alfonso, Chirico, Pantano, Zaccolo Improvement of a FRET-based indicator for cAMP by linker design and stabilization of donor-acceptor interaction. J Mol Bio. 2005;354:546–55. doi: 10.1016/j.jmb.2005.09.089. [DOI] [PubMed] [Google Scholar]
- Lissandron, Rossetto, Erbguth, Fiala, Daga, Zaccolo Transgenic fruit-flies expressing a FRET-based sensor for in vivo imaging of cAMP dynamics. Cellular Signalling. 2007;19:2296–303. doi: 10.1016/j.cellsig.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Liu, Zhang, Xiang FRET-based direct detection of dynamic protein kinase A activity on the sarcoplasmic reticulum in cardiomyocytes. Biochemical and biophysical research communications. 2011;404:581–6. doi: 10.1016/j.bbrc.2010.11.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohse, Nuber, Hoffmann Fluorescence/bioluminescence resonance energy transfer techniques to study G-protein-coupled receptor activation and signaling. Pharmacological Reviews. 2012;64:299–336. doi: 10.1124/pr.110.004309. [DOI] [PubMed] [Google Scholar]
- Macurek, Lindqvist, Lim, Lampson, Klompmaker, Freire, Clouin, Taylor, Yaffe, Medema Polo-like kinase 1 is activated by aurora A to promote checkpoint recovery. Nature. 2008;455:119–123. doi: 10.1038/nature07185. [DOI] [PubMed] [Google Scholar]
- Marvin, Borghuis, Tian, Cichon, Harnett, Akerboom, Gordus, Renninger, Chen, Bargmann, Orger, Schreiter, Demb, Gan, Hires, Looger An optimized fluorescent probe for visualizing glutamate neurotransmission. Nature Methods. 2013;10:162–70. doi: 10.1038/nmeth.2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta, Zhang Reporting from the field: genetically encoded fluorescent reporters uncover signaling dynamics in living biological systems. Ann Rev Biochem. 2011;80:375–401. doi: 10.1146/annurev-biochem-060409-093259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertens, Piljic, Schultz, Svergun Conformational analysis of a genetically encoded FRET biosensor by SAXS. Biophys J. 2012;102:2866–75. doi: 10.1016/j.bpj.2012.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mironov, Skorova, Taschenberger, Hartelt, Nikolaev, Lohse, Kugler Imaging cytoplasmic cAMP in mouse brainstem neurons. BMC Neurosci. 2009;10:29. doi: 10.1186/1471-2202-10-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyawaki Visualization of the spatial and temporal dynamics of intracellular signaling. Dev Cell. 2003;4:295–305. doi: 10.1016/s1534-5807(03)00060-1. [DOI] [PubMed] [Google Scholar]
- Mower, Kwok, Yu, Majewska, Okamoto, Hayashi, Sur Experience-dependent regulation of CaMKII activity within single visual cortex synapses in vivo. PNAS. 2011;108:21241–6. doi: 10.1073/pnas.1108261109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai, Yamada, Tominaga, Ichikawa, Miyawaki Expanded dynamic range of fluorescent indicators for Ca2+ by circularly permuted yellow fluorescent proteins. PNAS. 2004;101:10554–9. doi: 10.1073/pnas.0400417101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakai, Ohkura, Imoto A high signal-to-noise Ca2+ probe composed of a single green fluorescent protein. Nature Biotech. 2001;19:137–41. doi: 10.1038/84397. [DOI] [PubMed] [Google Scholar]
- Neininger, Thielemann, Gaestel FRET-based detection of different conformations of MK2. EMBO Reports. 2001;2:703–8. doi: 10.1093/embo-reports/kve157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, Zhang Visualization of phosphatase activity in living cells with a FRET-based calcineurin activity sensor. Molecular Biosystems. 2008;4:496–501. doi: 10.1039/b720034j. [DOI] [PubMed] [Google Scholar]
- Nikolaev, Moshkov, Lyon, Miragoli, Novak, Paur, Lohse, Korchev, Harding, Gorelik Beta2-adrenergic receptor redistribution in heart failure changes cAMP compartmentation. Science. 2010;327:1653–7. doi: 10.1126/science.1185988. [DOI] [PubMed] [Google Scholar]
- Nunes, Sample, Xiang, Monteiro, Gauda, Zhang Effect of oxygen on phosphodiesterases (PDE) 3 and 4 isoforms and PKA activity in the superior cervical ganglia. Adv Exp Med Biol. 2012;758:287–94. doi: 10.1007/978-94-007-4584-1_39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveria, Dittmer, Youn, Dell'Acqua, Sather Localized calcineurin confers Ca2+-dependent inactivation on neuronal L-type Ca2+ channels. J Neurosci. 2012;32:15328–37. doi: 10.1523/JNEUROSCI.2302-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira, Yasuda An improved Ras sensor for highly sensitive and quantitative FRET-FLIM imaging. PLoS One. 2013;8:e52874. doi: 10.1371/journal.pone.0052874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson, Liao, Sun, Roberts mRNA display selection of a high-affinity, modification-specific phospho-IkBa-binding fibronectin. ACS Chem Biol. 2008;3:480–5. doi: 10.1021/cb800069c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouyang, Sun, Chien, Wang Determination of hierarchical relationship of Src and Rac at subcellular locations with FRET biosensors. 2008. [DOI] [PMC free article] [PubMed]
- Padilla-Parra, Tramier FRET microscopy in the living cell: Different approaches, strengths and weaknesses. Bioessays. 2012;34:369–76. doi: 10.1002/bies.201100086. [DOI] [PubMed] [Google Scholar]
- Palmer, Giacomello, Kortemme, Hires, Lev-Ram, Baker, Tsien Ca2+ indicators based on computationally redesigned calmodulin-peptide pairs. Chemistry and Biology. 2006;13:521–530. doi: 10.1016/j.chembiol.2006.03.007. [DOI] [PubMed] [Google Scholar]
- Palmer, Qin, Park, McCombs Design and application of genetically encoded biosensors. Trends Biotechnol. 2011;29:144–52. doi: 10.1016/j.tibtech.2010.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrini, Camonis, Matsuda, de Gunzburg Dissecting activation of the PAK1 kinase at protrusions in living cells. JBC. 2009;284:24133–43. doi: 10.1074/jbc.M109.015271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Periasamy, Wallrabe, Chen, Barroso Quantitation of protein-protein interactions: confocal FRET microscopy. Methods in Cell Biol. 2008;89:569. doi: 10.1016/S0091-679X(08)00622-5. [DOI] [PubMed] [Google Scholar]
- Piljic, Diego, Wilmanns, Schultz Rapid development of genetically encoded FRET reporters. ACS Chem Biol. 2011;6:685–91. doi: 10.1021/cb100402n. [DOI] [PubMed] [Google Scholar]
- Piston, Rizzo FRET by fluorescence polarization microscopy. Methods in Cell Biology. 2008;85:415–30. doi: 10.1016/S0091-679X(08)85018-2. [DOI] [PubMed] [Google Scholar]
- Ran, Hsu, Wright, Agarwala, Scott, Zhang Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rebois, Robitaille, Petrin, Zylbergold, Trieu, Hebert Combining protein complementation assays with resonance energy transfer to detect multipartner protein complexes in living cells. Methods. 2008;45:214–8. doi: 10.1016/j.ymeth.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Rothman, Shults, Imperiali Chemical approaches for investigating phosphorylation in signal transduction networks. Trends in Cell Bio. 2005;15:502–10. doi: 10.1016/j.tcb.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Sanderson, Dell'Acqua AKAP signaling complexes in regulation of excitatory synaptic plasticity. The Neuroscientist. 2011;17:321–36. doi: 10.1177/1073858410384740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki, Sato, Umezawa Fluorescent indicators for Akt/protein kinase B and dynamics of Akt activity visualized in living cells. J Biol Chem. 2003;278:30945–30951. doi: 10.1074/jbc.M212167200. [DOI] [PubMed] [Google Scholar]
- Sato, Umezawa Imaging protein phosphorylation by fluorescence in single living cells. Methods. 2004;32:451–5. doi: 10.1016/j.ymeth.2003.10.013. [DOI] [PubMed] [Google Scholar]
- Sato, Kawai, Umezawa Genetically encoded fluorescent indicators to visualize protein phosphorylation by extracellular signal-related kinase in single living cells. Anal Chem. 2007;79:2570–5. doi: 10.1021/ac062171d. [DOI] [PubMed] [Google Scholar]
- Sellers, De Arcangelis, Xiang, Best Cardiomyocytes with disrupted CFTR function require CaMKII and Ca(2+)-activated Cl(−) channel activity to maintain contraction rate. J Physiol. 2010;588:2417–29. doi: 10.1113/jphysiol.2010.188334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seong, Ouyang, Kim, Sun, Wen, Lu, Zhou, Llewellyn, Schlaepfer, Guan, Chien, Wang Detection of focal adhesion kinase activation at membrane microdomains by fluorescence resonance energy transfer. Nature Comm. 2011;2 doi: 10.1038/ncomms1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelly, Lim, Cancedda, Heilshorn, Gao, Poo Local and long-range reciprocal regulation of cAMP and cGMP in axon/dendrite formation. Science. 2010;327:547–52. doi: 10.1126/science.1179735. [DOI] [PubMed] [Google Scholar]
- Shelly, Lim, Cancedda, Popescu, Cheng, Gao, Poo Semaphorin3A regulates neuronal polarization by suppressing axon formation and promoting dendrite growth. Neuron. 2011;71:433–46. doi: 10.1016/j.neuron.2011.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sin, Edwards, Li, Day, Christian, Dunlop, Adams, Zaccolo, Houslay, Baillie Disruption of the cyclic AMP phosphodiesterase-4 (PDE4)-HSP20 complex attenuates the beta-agonist induced hypertrophic response in cardiac myocytes. J Mol Cell Cardiol. 2011;50:872–83. doi: 10.1016/j.yjmcc.2011.02.006. [DOI] [PubMed] [Google Scholar]
- Sipieter, Vandame, Spriet, Leray, Vincent, Trinel, Bodart, Riquet, Heliot . From FRET Imaging to Practical Methodology for Kinase Activity Sensing in Living Cells. In: Morris MC, editor. Fluorescence-Based Biosensors: From Concepts To Applications. Elsevier Academic Press; San Diego, CA: 2013. pp. 145–216. [DOI] [PubMed] [Google Scholar]
- Song, Rodgers, Schultz, Liao Protein interaction affinity determination by quantitative FRET technology. Biotech Bioeng. 2012;109:2875. doi: 10.1002/bit.24564. [DOI] [PubMed] [Google Scholar]
- Spencer, Cappell, Tsai, Overton, Wang, Meyer The proliferation-quiescence decision is controlled by a bifurcation in CDK2 activity at mitotic exit. Cell. 2013;155:369–83. doi: 10.1016/j.cell.2013.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Rombola, Jyothikumar, Periasamy Forster resonance energy transfer microscopy and spectroscopy for localizing protein-protein interactions in living cells. Cytometry A. 2013;83:780–93. doi: 10.1002/cyto.a.22321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao, Okamoto, Nakagawa, Neve, Nagai, Miyawaki, Hashikawa, Kobayashi, Hayashi Visualization of synaptic Ca2+/calmodulin-dependent protein kinase II activity in living neurons. J Neurosci. 2005;25:3107–12. doi: 10.1523/JNEUROSCI.0085-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tantama, Hung, Yellen Optogenetic reporters: Fluorescent protein-based genetically encoded indicators of signaling and metabolism in the brain. Prog Brain Res. 2012;196:235–63. doi: 10.1016/B978-0-444-59426-6.00012-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai, Matsuda Ras binding opens c-Raf to expose the docking site for mitogen-activated protein kinase kinase. EMBO Rep. 2005;6:251–5. doi: 10.1038/sj.embor.7400349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terai, Matsuda The amino-terminal B-raf-specific region mediates calcium-dependnt homo- and hetero-dimerization of Raf. EMBO J. 2006;25:3556–64. doi: 10.1038/sj.emboj.7601241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timm, von Kries, Li, Zempel, Mandelkow, Mandelkow Microtubule affinity regulating kinase activity in living neurons was examined by a genetically encoded fluorescence resonance energy transfer/fluorescence lifetime imaging-based biosensor: inhibitors with therapeutic potential. J Biol Chem. 2011;286:41711–22. doi: 10.1074/jbc.M111.257865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting, Kain, Klemke, Tsien Genetically encoded fluorescent reporters for tyrosine kinase activities in living cells. PNAS. 2001;98:15003–8. doi: 10.1073/pnas.211564598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomida, Takekawa, O'Grady, Saito Stimulus-specific distinctions in spatial and temporal dynamics of stress-activated protein kinase kinase kinases revealed by a fluorescence resonance energy transfer biosensor. Mol Cell Biol. 2009;29:6117–27. doi: 10.1128/MCB.00571-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsalkova, Mei, Li, Chepurny, Leech, Liu, Holz, Woods, Cheng Isoform-specific antagonists of exchange proteins directly activated by cAMP. PNAS. 2012;109:18613–8. doi: 10.1073/pnas.1210209109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou, Zheng, Hsu, Sasaki, Cantley A fluorescent reporter of AMPK activity and cellular energy stress. Cell Metab. 2011;13:476–86. doi: 10.1016/j.cmet.2011.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaasa, Lust, Terrin, Uri, Zaccolo Small-molecule FRET probes for protein kinase activity monitoring in living cells. Biochem Biophys Res Comm. 2010;397:750–5. doi: 10.1016/j.bbrc.2010.06.026. [DOI] [PubMed] [Google Scholar]
- Vincent, Castro, Gervasi, Guiot, Brito, Paupardin-Tritsch PDE4 Control on cAMP/PKA Compartmentation Revealed by Biosensor Imaging in Neurons. Horm Metab Res. 2010;44:786–9. doi: 10.1055/s-0032-1311631. [DOI] [PubMed] [Google Scholar]
- Violin, Zhang, Tsien, Newton A genetically encoded fluorescent reporter reveals oscillatory phosphorylation by protein kinase C. J Cell Biol. 2003;281:30947–56. doi: 10.1083/jcb.200302125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt, Schmidt, Lohse, Hashimoto, Bock, Kudia Multicolor bimolecular fluorescence complementation reveals simultaneous formation of alternative CBL/CIPK complexes in planta. Plant J. 2008;56:505–16. doi: 10.1111/j.1365-313X.2008.03612.x. [DOI] [PubMed] [Google Scholar]
- Wang, Botvinick, Zhao, Berns, Usami, Tsien, Chien Visualizing the mechanical activation of Src. Nature. 2005;434:1040–5. doi: 10.1038/nature03469. [DOI] [PubMed] [Google Scholar]
- Wang, Govindaiah, Liu, De Arcangelis, Cox, Xiang Binding of amyloid beta peptide to beta2 adrenergic receptor induces PKA-dependent AMPA receptor hyperactivity. FASEB J. 2010;24:3511–21. doi: 10.1096/fj.10-156661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werthmann, Volpe, Lohse, Calebiro Persistent cAMP signaling by internalized TSH receptors occurs in thyroid but not in HEK293 cells. FASEB J. 2012;26:2043. doi: 10.1096/fj.11-195248. [DOI] [PubMed] [Google Scholar]
- Williams, Fogarty, Tsien, Fay Calcium gradients in single smooth-muscle cells revealed by the digital imaging microscope using fura-2. Nature. 1985;318:558–61. doi: 10.1038/318558a0. [DOI] [PubMed] [Google Scholar]
- Willoughby, Masada, Wachten, Pagano, Halls, Everett, Ciruela, Cooper AKAP79/150 interacts with AC8 and regulates Ca2+-dependent cAMP synthesis in pancreatic and neuronal systems. J Biol Chem. 2010;285:20328–42. doi: 10.1074/jbc.M110.120725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woeher, Wlodarczyk, Neher Signal/noise analysis of FRET-based sensors. Biophys J. 2010;99:2344. doi: 10.1016/j.bpj.2010.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu, Hudry, Hashimoto, Uemura, Fan, Berezovska, Grosskreutz, Bacskai, Hyman Distinct dendritic spine and nuclear phases of calcineurin activation after exposure to amyloid-beta revealed by a novel fluorescence resonance energy transfer assay. J Neurosci. 2012;32:5298–309. doi: 10.1523/JNEUROSCI.0227-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu-Zhang, Murphy, Bachman, Newton Isozyme-specific interaction of protein kinase C delta with mitochondria dissected using live cell fluorescence imaging. J Biol Chem. 2012;287:37891–906. doi: 10.1074/jbc.M112.412635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Peter, Bouquier, Ollendorff, Fillamil, Liu, Fagni, Perroy REV, a BRET-based sensor of Erk activity. Front Endocrinol. 2013;4:95. doi: 10.3389/fendo.2013.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, Shinotsuka, Nonomura, Takemoto, Kuida, Yosida, Miura Live imaging of apoptosis in a novel transgenic mouse highlights its role in neural tube closure. J Cell Bio. 2011;195:1047–60. doi: 10.1083/jcb.201104057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda Studying Signal Transduction in Single Dendritic Spines. Cold Spring Harbor Perspectives in Biology. 2012;4:a005611. doi: 10.1101/cshperspect.a005611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda, Murakoshi The mechanisms underlying the spatial spreading of signaling activity. Curr Op Neurobio. 2011;21:313–21. doi: 10.1016/j.conb.2011.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshizaki, Mochizuki, Gotoh, Matsuda Akt-PDK1 complex mediates epidermal growth factor-induced membrane protrusion through Ral activation. Mol Biol Cell. 2007;18:119–28. doi: 10.1091/mbc.E06-05-0467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaccolo, De Giorgi, Cho, Feng, Knapp, Negulescu, Taylor, Tsien, Pozzan A genetically encoded fluorescent indicator for cyclic AMP in living cells. Nature Cell Biol. 2000;2:25–9. doi: 10.1038/71345. [DOI] [PubMed] [Google Scholar]
- Zaccolo, Pozzan Discrete microdomains with high concentration of cAMP in stimulated rat neonatal cardiac myocytes. Science. 2002;295:1711–15. doi: 10.1126/science.1069982. [DOI] [PubMed] [Google Scholar]
- Zadran, Jourdi, Rostamiani, Qin, Bi, Baudry Brain-derived neurotrophic factor and epidermal growth factor activate neuronal m-calpain via mitogen-activated protein kinase-dependent phosphorylation. J Neurosci. 2010;30:1086–95. doi: 10.1523/JNEUROSCI.5120-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Ma, Taylor, Tsien Genetically Encoded Reporters of Protein Kinase A activity Reveal Impact of Substrate Tethering. PNAS. 2001;98:14997–15002. doi: 10.1073/pnas.211566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Lee, Bhojani, Khan, Shilman, Holland, Ross, Rehemtulla Molecular imaging of Akt kinase activity. Nature Medicine. 2007;13:1114–9. doi: 10.1038/nm1608. [DOI] [PubMed] [Google Scholar]
- Zhou, Gao, Han, Brinker, Caldwell, Gu An intracellular conformational sensor assay for Abl T315I. Analytical Biochem. 2009;385:300–8. doi: 10.1016/j.ab.2008.11.015. [DOI] [PubMed] [Google Scholar]