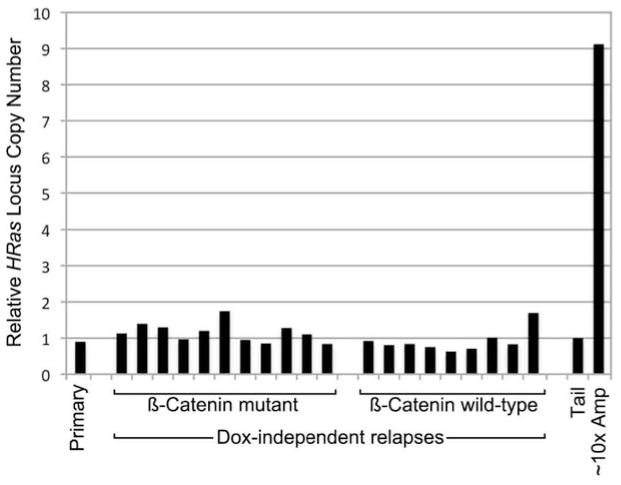
Extended Data Figure 8. Increased HRas MAFs in βcatmut DITs is not due to gross copy changes at the HRas locus.
Histogram depicts HRas allele copy number relative to β-actin determined for a cohort of clonally-related Wnt tumor outgrowths. Independent relapse samples are presented in the same order depicted in Fig. 4b. Copy number values were obtained by performing qPCR on genomic DNA from tumor samples and from normal tail, with tail values set at 1. As a positive control, we included a p19Arf-deficient Wnt tumor sample (~10x Amp) previously found to have approximately 10-fold HRas copy number gain as determined by Southern hybridization. Since, HRas MAFs reproducibly exceeded βcat MAFs by approximately 2-fold across the βcatmut relapse set (Fig. 4b), elevated HRas MAFs may reflect duplication of the HRasmut allele (e.g., via a gene conversion event) sometime in the life history of βcatmut subclones.