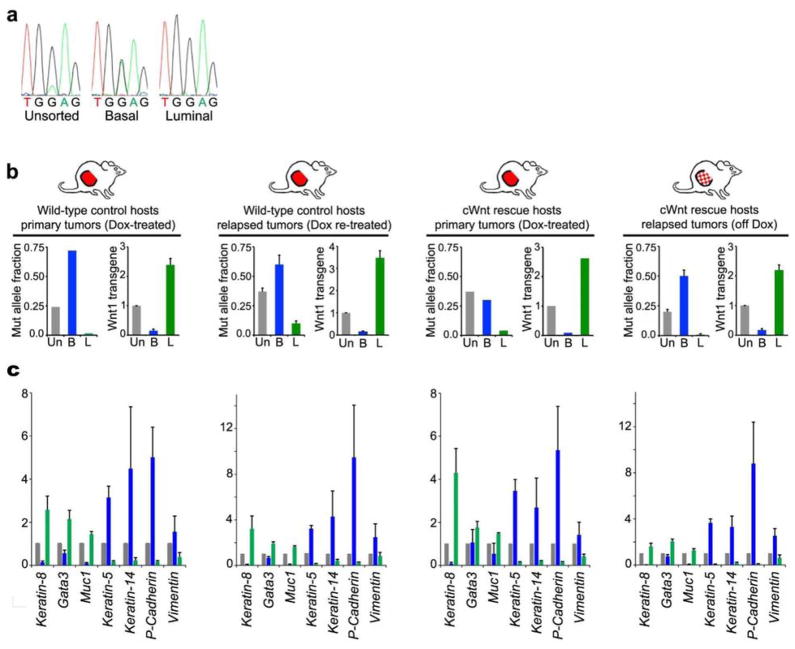
Extended Data Figure 4. Biclonal configuration of reconstituted iWnt/mRFP+ tumors.
a, DNA sequencing chromatograms depicting a basally-enriched HRasGGA12GAA mutation detected in the parental tumor. b, Evidence for distinct basal HRasmut/Wnt1low and luminal HRaswt/Wnt1high tumor subclones. Sorted tumor cell subsets were analyzed by DNA sequencing and by qRT-PCR for Wnt1 expression relative to Gapdh. Histograms at left show HRas MAFs determined from chromatogram peak heights. Histograms at right show relative Wnt1 expression with values from unsorted tumor cells set at 1. Un, unsorted; B, basal; L, luminal. Data represent mean +/− SEM for (from left to right) n = 2, 4, 3, 4, 1, 2, 6, or 12 explants. c, For each condition, sorted tumor cell subsets were analyzed by qRT-PCR for expression of several epithelial lineage-specific genes relative to Gapdh, with values for unsorted tumor cells set at 1. Gray bars, unsorted; Blue bars, basal; Green bars, luminal. Data represent mean +/− SEM for (from left to right) n = 4, 4, 3, or 12 explants.