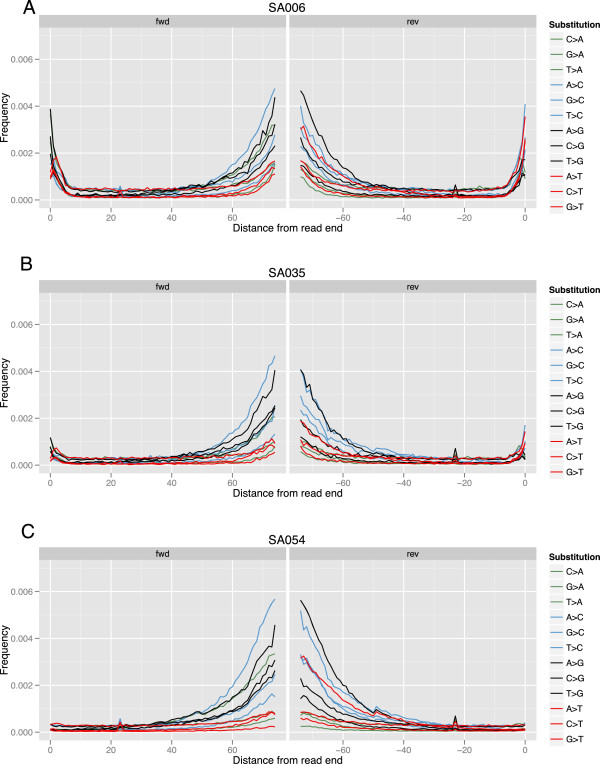
Figure 2.
Assessment of base substitutions from mapped reads. Each mapped read was compared to the genome reference sequence to assess patterns consistent with DNA degradation. At each of the 75 positions along a read, we plot the frequency of substitution types, for both the forward (left) and reverse (right) reads from each read-pair. Analysis was limited to 1 million reads from chromosome 1; all raw reads are plotted. Three individuals with varying levels of substitution errors are shown: (A) SA006 with overall higher substitution rate and an excess of purines at the start of the first read, (B) SA035 with a slightly elevated substitution rate and excess of purines at the start of the first read, and (C) SA054 with a low substitution rate and no bias at the beginning of the first read. The additional five Pilot 1 individuals tended to resemble SA054 (Additional file 1: Figure S4). Removal of reads with any soft-clipping substantively reduced the mis-incorporation rate for SA006 and SA035.