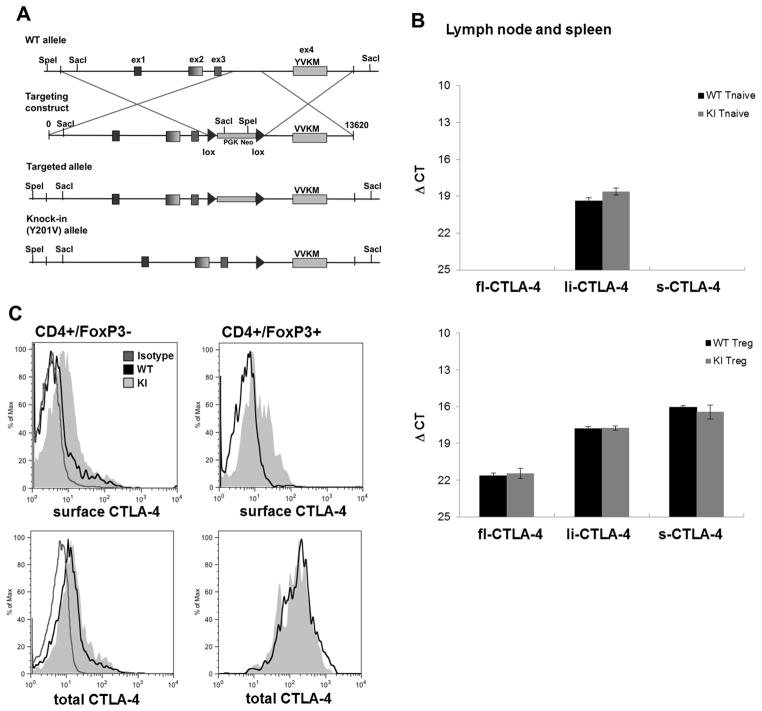
Figure 1.
(A) A 13.6 kilobase genomic fragment containing the entire mouse CTLA-4 locus was retrieved from a bacterial artificial chromosome (clone RP23-146J17: BACPAC). The nucleotide sequence was modified resulting in an amino acid change from Tyrosine (Y) to Valine (V) at position 201 within Exon4. Further, a LoxP-flanked PGK/em7-promotor driven neo cassette was inserted to allow selection of successful targeted B6 ES cell clones. Deletion of the NEO-cassette was achieved by breeding founder mice with OX40-Cre transgenic mice. (B) T naïve and T regulatory cells of CTLA-4 WT and Y201V knock-in mice were FACS sorted and mRNA expression levels of the different CTLA-4 isoforms were measured by quantitative real-time PCR. The ΔCt (threshold cycle) value was determined using the following formula: . Data are displayed as mean ΔCt ± SD. (C) Surface (upper panel) and total (lower panel) CTLA-4 expression in primary CD4+/FoxP3− T conventional and CD4+ FoxP3+ T regulatory cells from WT and Y201V knock-in mice was assessed by flow cytometry.