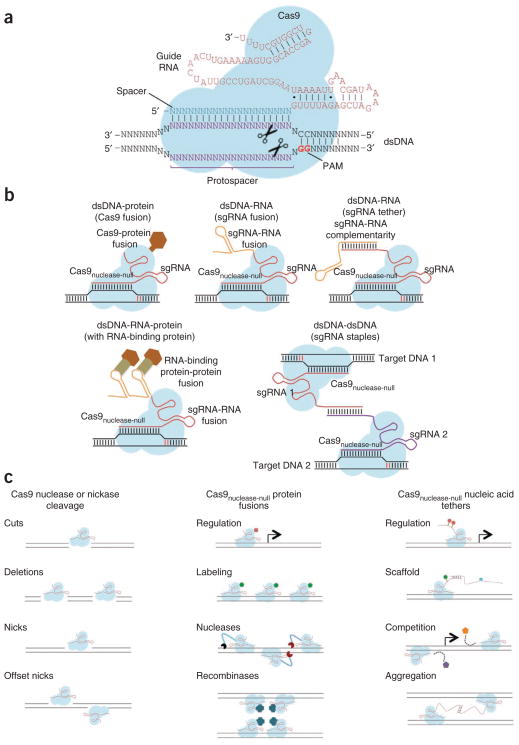
Figure 2.
Cas9-sgRNA targeting complexes. (a) The basic S. pyogenes Cas9-sgRNA RNA-guided nuclease complex for eukaryotic genome engineering. Target recognition and cleavage require protospacer sequence complementary to the spacer and presence of the appropriate NGG PAM sequence at the 3′ of the protospacer. (b) Cas9 enables programmable localization of dsDNA, RNA and proteins. Proteins can be targeted to any dsDNA sequence by simply fusing them to Cas9nuclease-null, and additional RNA can be tethered to sgRNA termini without compromising Cas9 binding. Attaching RNA binding sites can in turn recruit RNA-binding proteins or directly recruit other RNAs via sequence hybridization. Finally, Cas9 can theoretically bring together any two dsDNA regions by employing sgRNA-Cas9 ‘staples’ that bind to the targeted loci and to one another. Consequently, Cas9 can in principle bring together any fusion proteins, any natural or fusion RNAs and/or any dsDNA locus to any other dsDNA sequence of interest. (c) The diverse potential applications of Cas9 range from targeted genome editing (via simplex and multiplex double-strand breaks and nicks) to targeted genome regulation (via tethering of epigenetic effector domains to either the Cas9 or sgRNA, and via competition with endogenous DNA binding factors) and possibly programmable genome reorganization and visualization. Cas9 might also be engineered to function as an RNA-guided recombinase, and via RNA tethers could serve as a scaffold for the assembly of multiprotein and nucleic acid complexes.