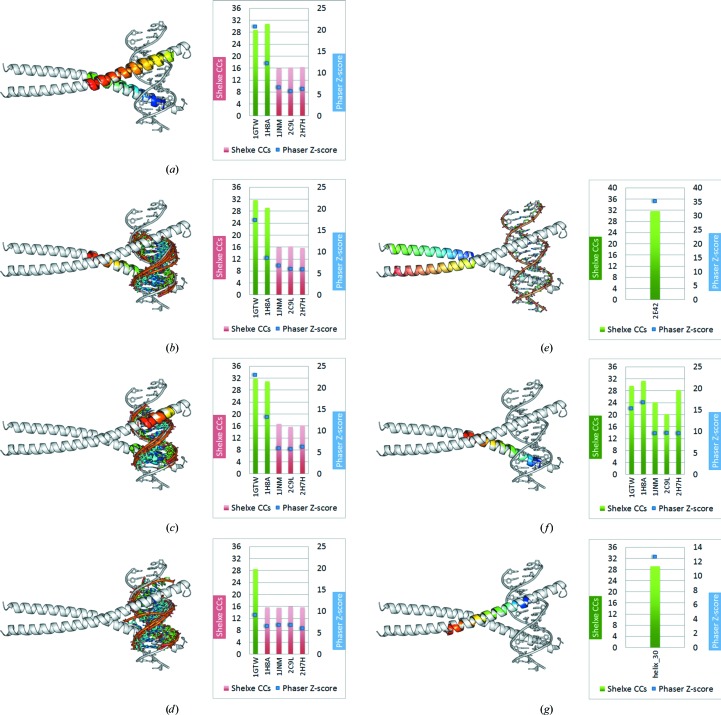
Figure 6.
Zipper-type target 2E42 with modified models as input to ARCIMBOLDO: (a) using only the helices (both) from the models leads to solutions for just two (1gtw and 1h8a) of the five fragments; (b) using as search fragments just one long helix (30 amino acids) and the DNA fragment leads to solutions in only two of the five models (1gtw and 1h8a); (c) the same two fragments (1gtw and 1h8a) also lead to a solution if the DNA plus shorter helices (12 amino acids each) are used as search fragments; (d) using only the DNA of the models as a search fragment leads to a solution in only one case (1gtw); (e) using the DNA-distant helices taken from the target structure 2E42 as search fragments leads to a clear solution; (f) cutting down this fragment even more to just one helix without the DNA leads to a solution for all five of the models (1gtw, 1h8a, 1jnm, 2c9l and 2h7h); (g) even searching for two copies of a model helix of 30 amino acids leads to a solution as the DNA-binding part of the zipper helix is quite straight and does not deviate much from an ideal straight model helix. The smallest solving fragment represents 8.13% of the mass of the asymmetric unit.