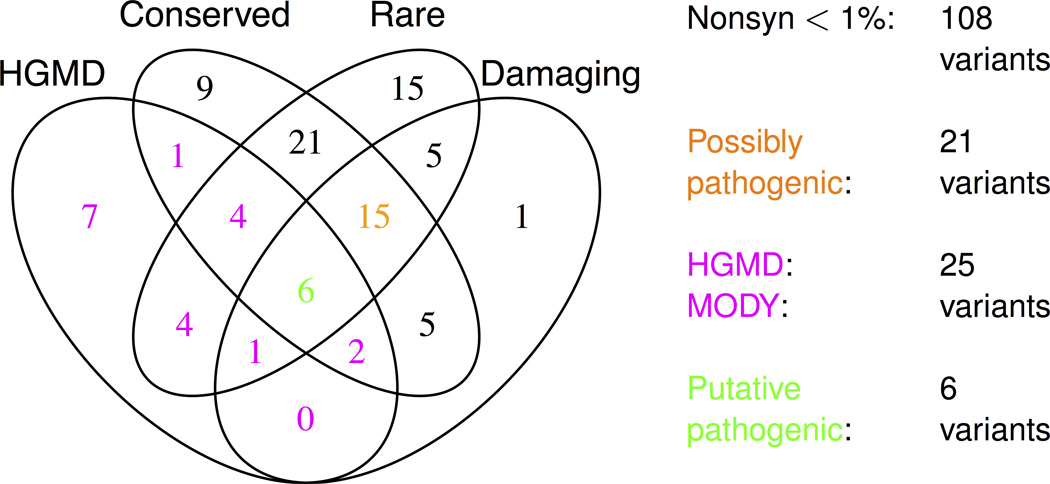
Figure 1. Description of low frequency non-synonymous variants.
Shown is a summary of all low frequency (MAF<1%) non-synonymous variants identified in the three cohorts (Supplementary Tables 1–3 show lists of all variants, not simply those of low frequency). MAF is calculated as the maximum frequency across the three cohorts. Shown also is the number of variants fitting each annotation (HGMD, conserved, rare, and damaging) and each variant class: low frequency non-synonymous (black, magenta, orange, or green), possibly pathogenic (orange or green), HGMD MODY (magenta or green), and putative pathogenic (green). Twelve low frequency variants fit no annotations. Low frequency non-synonymous is abbreviated to ‘Nonsyn <1%’.