Fig. 7.
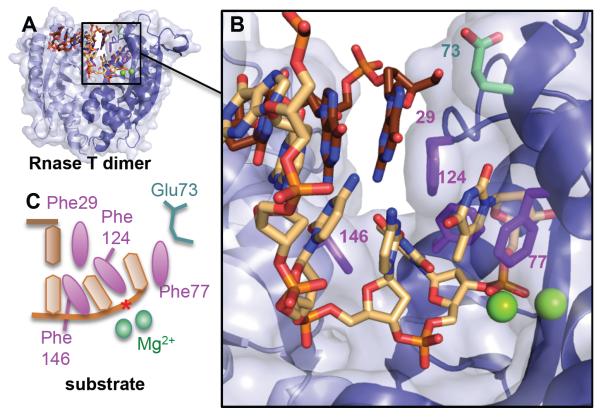
Rnase T dimer selects for ssDNA or ssRNA or 3′ overhangs by blocking the complementary strand (brown) with a steric helical wedge and Phe29. A) Global and B) closeup of the duplex structure (3va3.pdb) with Mg2+ atoms from an overlaid RnaseT/ssDNA structure (3v9w.pdb) to show relative position of active site metals and scissile phosphate (*). C) DNA schematic showing how the 3′ overhang is then selected through intercalation by Phe146, Phe124, and Phe77 and positioning of the phosphodiester bond for catalysis. For terminal cytosines with reduced incision activity, Glu73 will flip and pull on the cytosine, disrupting the positioning of the phosphodiester backbone and one of the two Mg2+ ions.