Figure 5.
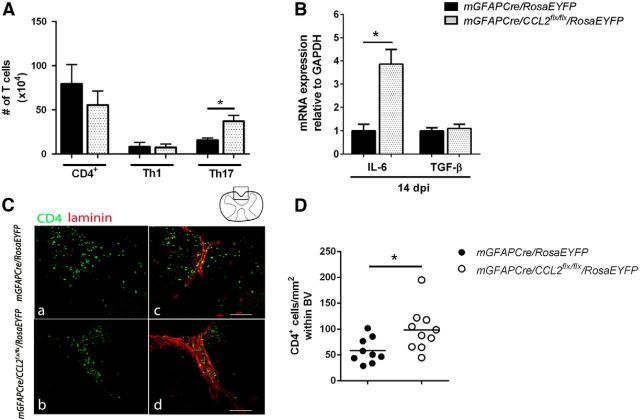
Flow cytometric analysis and spatial distribution of CNS CD4 T cells. A, Mononuclear cells were isolated from pooled brain/spinal cord and analyzed by flow cytometry at 16 dpi. Total CD4 (CD4+), and Th1 (IFNγ+IL-17−), did not show statistically significant changes between the two groups of mice. The number of Th17 (IFNγ−IL-17+) cells were upregulated in the CNS of mGFAPCre/CCL2flx/flx/RosaEYFP mice compared with controls. B, IL-6 and TGF-β RNAs were quantified using real-time RT-PCR of lumbar spinal cord RNA extracts at 16 dpi. IL-6 mRNA was increased, but TGF-β mRNA was unchanged, in mGFAPCre/CCL2flx/flx/RosaEYFP mice compared with littermate controls. n = 6 for mGFAPCre/RosaEYFP; n = 8 for mGFAPCre/CCL2flx/flx/RosaEYFP. *p = 0.0119 (unpaired t test). C, Retention of CD4+ T cells (green) within the perivascular space of lumbar spinal cord shown by laminin (red) in mGFAPCre/CCL2flx/flx/RosaEYFP mice compared with controls. D, Quantification of CD4+ T cells observed within spinal cord ventral blood vessels was determined using ImageJ analysis (triplicate measurements were performed for each mouse). *p = 0.0229.