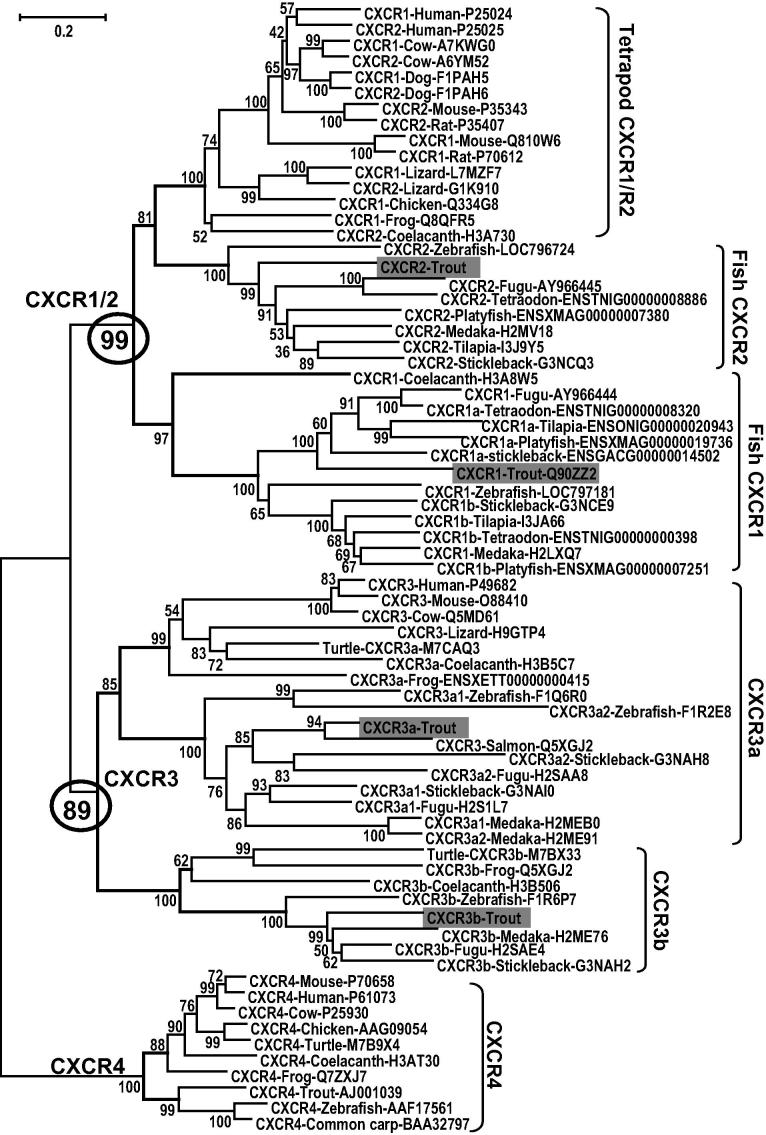
Fig. 3.
An unrooted phylogenetic tree of vertebrate CXCR1–4 molecules. The tree was constructed using amino acid multiple alignments and the neighbour-joining method within the MEGA5 program (Tamura et al., 2011). Node values represent percent bootstrap confidence derived from 10,000 replicates. The evolutionary distances were computed using the JTT matrix-based method. All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons. The accession number for each sequence is given after the molecular type and species name except for trout CXCR2, CXCR3a and CXCR3b that are provided in Table 2.