Abstract
Gamma-aminobutyric acid (GABA), a building block of the biodegradable plastic polyamide 4, is synthesized from glucose by Corynebacterium glutamicum that expresses Escherichia coli glutamate decarboxylase (GAD) B encoded by gadB. This strain was engineered to produce GABA more efficiently from biomass-derived sugars. To enhance GABA production further by increasing the intracellular concentration of its precursor glutamate, we focused on engineering pknG (encoding serine/threonine protein kinase G), which controls the activity of 2-oxoglutarate dehydrogenase (Odh) in the tricarboxylic acid cycle branch point leading to glutamate synthesis. We succeeded in expressing GadB in a C. glutamicum strain harboring a deletion of pknG. C. glutamicum strains GAD and GAD ∆pknG were cultured in GP2 medium containing 100 g L−1 glucose and 0.1 mM pyridoxal 5′-phosphate. Strain GAD∆pknG produced 31.1 ± 0.41 g L−1 (0.259 g L−1 h−1) of GABA in 120 hours, representing a 2.29-fold higher level compared with GAD. The production yield of GABA from glucose by GAD∆pknG reached 0.893 mol mol−1.
Keywords: Corynebacterium glutamicum, Gamma-aminobutyric acid, Glutamate decarboxylase, 2-oxoglutarate dehydrogenase, Protein kinase G
Introduction
Diverse microorganisms, animals, and plants synthesize the amino acid gamma-aminobutyric acid (GABA), which does not naturally occur in proteins. GABA functions as a neurotransmitter in humans, lowers blood pressure (Hayakawa et al. [2004]), and is a component of pharmaceuticals and foods (Li and Cao [2010]). The bioplastic polyamide 4 (PA4) is a linear polymer of GABA, which is chemically synthesized from the GABA lactam 2-pyrrolidone (Kawasaki et al. [2005]). PA4 has excellent physical properties based on its high melting point of 260°C and its degradability by microbes in soil (Hashimoto et al. [1994]) and activated sludge (Yamano et al. [2008]). The synthesis of GABA from abundantly available biomass by recombinant microorganisms will make it possible to produce new bioplastics at low cost.
Glutamate decarboxylase (GAD; EC 4.1.1.15) catalyzes the conversion of L-glutamate to GABA through alpha-decarboxylation (Fonda [1985]). Genes (gad) encoding GAD are present in Escherichia coli (DeBiase et al. [1996]), Lactobacillus brevis (Oda et al. [2008]), Lactobacillus paracasei (Shima et al. [2008]), and several other species of Lactobacillus as well as Enterobacteria. Lactic acid bacteria produce GABA when glutamate is added to the fermentation medium. Although the quantities of GABA produced by this method are sufficient for producing foods, it is not cost-effective for producing chemicals.
Therefore, we developed a robust system for producing GABA from saccharides by expressing E. coli GAD in Corynebacterium glutamicum (Takahashi et al. [2012]). The biotin auxotroph C. glutamicum is a nonpathogenic, nonsporulating, nonmotile, Gram-positive soil bacterium that belongs to the order Actinomycetales, which includes Corynebacteria, Nocardia, Rhodococci, and other related microorganisms (George [2001]). C. glutamicum is an important industrial microorganism, because it produces high levels of glutamate and other amino acids, which are widely used in pharmaceuticals, animal feed, and food supplements (Leuchtenberger et al. [2005], Hermann [2003]). To efficiently produce GABA, the gadB gene from E. coli W3110 was overexpressed in a glutamate-producing C. glutamicum strain (ATCC 13032). After optimization, this strain produced 12.37 g L−1 of GABA from glucose in the presence of pyridoxal 5′-phosphate in the absence of added glutamate (Takahashi et al. [2012]).
In the present study, we further enhanced GABA synthesis using recombinant C. glutamicum strains expressing GAD to optimize the intracellular level of glutamate. To increase the flux of 2-oxoglutarate to glutamate, we attempted to disrupt pknG, which affects the activity of 2-oxoglutarate dehydrogenase (Odh). The reduction of the 2-oxoglutarate dehydrogenase complex (ODHC) is an important factor for glutamate synthesis by C. glutamicum (Kimura [2002]). ODHC participates in the tricarboxylic acid (TCA) cycle and catalyzes the conversion of 2-oxoglutarate to succinyl-CoA. The ODHC comprises four subunits, OdhI, OdhA, AceF, and Lpd. The activity of C. glutamicum ODHC is controlled by a regulatory mechanism that involves OdhI and serine/threonine protein kinase G (PknG, EC 2.7.11.1) (Schultz et al. [2009]). PknG catalyzes the phosphorylation of OdhI, a 15 kDa subunit of ODHC. Unphosphorylated OdhI binds the EI subunit (OdhA) of ODHC and inhibits its activity. Inhibition of ODHC activity is reversed by phosphorylation of OdhI at threonine residue 14 by PknG (Niebisch et al. [2006]). The pknG-deficient mutant produces glutamate at a higher rate compared with the parental C. glutamicum strain, suggesting that the mutations influence ODHC activities (Schultz et al. [2007], Boulahya et al. [2010]).
In the present study, a pknG-deficient C. glutamicum strain expressing GAD was generated to increase the flux of 2-oxoglutarate towards glutamate for more efficient biosynthesis of GABA. Using this strain, we were able to produce significantly higher levels of GABA from glucose.
Materials and methods
Bacterial strains and media
The bacterial strains and plasmids used in this study are listed in Table 1. E. coli strains were grown in Luria-Bertani medium (10 g L−1 tryptone, 5 g L−1 yeast extract, and 5 g L−1 sodium chloride) containing 50 μg mL−1 kanamycin at 37°C. C. glutamicum ATCC 13032 and all recombinant strains were grown in brain–heart infusion (BHI) medium (Becton, Dickinson and Co., Franklin Lakes, NJ, USA). BHI medium supplemented with 25 μg mL−1 kanamycin and 1.5% agar was used to select C. glutamicum transformants. The transformants were first cultivated at 30°C for 24 hours in a test tube containing 5 mL BHI medium with 25 μg mL−1 kanamycin and then inoculated into 20 mL GP2 medium (Takahashi et al. [2012]) containing 25 μg mL−1 kanamycin in a 200-mL flask for fermentation.
Table 1.
Bacterial strains and plasmids
| Bacterial strains or plasmids | Relevant characteristics | Reference or source |
|---|---|---|
|
E. coli |
|
|
| SCS110 |
rpsL (Strr) thr leu endA thi-l lacY galK galT ara tonA tsx dam dcm |
Stratagene |
| |
supE44Δ (lac-proAB) [F’traD36 proAB laclqZΔM15] |
|
|
C. glutamicum |
|
|
| ATCC 13032 |
Wild-type C. glutamicum, biotin-auxotrophic, L-glutamate producing strain |
ATCC |
| W |
Wild-type C. glutamicum derivative harboring pCH |
Takahashi et al. [2012] |
| GAD |
Wild-type C. glutamicum derivative harboring pCH-gadB |
Takahashi et al. [2012] |
| ΔpknG |
Wild-type C. glutamicum derivative with deletion in pknG |
This study |
| GADΔpknG |
Wild-type C. glutamicum derivative with deletion in pknG, harboring pCH-gadB |
This study |
| Plasmids |
|
|
| pCH |
E. coli-C. glutamicum shuttle vector with HCE promoter, Kmr |
Tateno et al. [2007] |
| pCH-gadB |
pCH containing gadB from E.coli W3110, Kmr |
Takahashi et al. [2012] |
| pTM44 |
pHSG298 with B. subtilis sacB and C. glutamicum hom, Kmr |
Mimitsuka et al. [2007] |
| pTM44-ΔpknG | pTM44 with 1,601-bp SphI-BamHI fragment with deletions of pknG, Kmr | This study |
r antibiotic resistance.
Molecular genetic techniques
E. coli SCS110 was used to avoid DNA methylation, and polymerase chain reaction (PCR) was conducted using KOD-Plus2 DNA polymerase (Toyobo, Osaka, Japan). Plasmid DNA was purified using a LaboPass™ Plasmid Mini Purification Kit (Cosmo Bio Co., Ltd., Tokyo, Japan).
Construction of plasmids for disrupting pknG
Genomic DNA was purified using the Wizard Genomic DNA Purification Kit (Promega, Madison, WI, USA) from C. glutamicum ATCC 13032 grown in BHI medium. The oligonucleotide primers used in this study are listed in Table 2. To inactivate pknG (KEGG Entry NCgl 2655, Gene Name Cgl 2751, 2469 nt), an 801-base pair (bp) upstream region of pknG (pknG-up) was amplified using PCR with the primer pair pknG-up-In-F and pknG-up-In-R-801 and strain ATCC 13032 genomic DNA as template. An 800-bp downstream region of pknG (pknG-down) was amplified using PCR with the primer pair pknG-up-In-F-1669 and pknG-down-In-R. The two fragments, pknG-up and pknG-down, were fused using overwrap PCR using the primer pair pknG-In-F2 and pknG-In-R2, yielding a 1,601-bp fragment of pknG. The amplified DNA fragment was purified from a 1.0% agarose gel using the Wizard SV Gel and PCR Clean-Up systems (Promega). The plasmid pTM44 (Mimitsuka et al. [2007]), which contains sacB from B. subtilis, was used as a suicide vector for markerless gene disruption and was digested with SphI and BamHI (New England BioLabs Inc., MA, USA) to remove a 1,344-bp SphI-BamHI fragment. The 1.60-kbp ΔpknG fragment was inserted into SphI-BamHI-digested pTM44 using an In Fusion HD Cloning Kit (Clontech Laboratories Inc., Mountain View, CA, USA), yielding pTM44-ΔpknG. The DNA sequences of the constructs were determined using an ABI PRISM 3100 Genetic Analyzer (Life Technologies, Carlsbad, CA, USA).
Table 2.
Oligonucleotide primers
| Primer name | Sequence (5′- 3′) | Restriction enzyme |
|---|---|---|
| pknG-up-In-F |
GCCAAGCTTgcatgcATGAAGGATAATGAAGATTTCGATCCAGATTCACCAGC |
SphI |
| pknG-up-In-R-801 |
ACCATTTGTGTCGCCGGCTTTGCAGCGGTCTTTCAGGGA |
|
| pknG-down-In-F-1669 |
GCCAAGCTTgcatgcGGCGACACAAATGGTTCTCCG |
SphI |
| pknG-down-In-R |
AAAAGGATCggatccCTAGAACCAACTCAGTGGCCGCA |
BamHI |
| pknG-In-F2 |
CCAGTGCCAAGCTTgcatgcATGAAGGATAATGAAGATTTCGATCCAGATTCACCAGC |
SphI |
| pknG-In-R2 |
AAAAAGGATCggatccCTAGAACCAACTCAGTGGCCGCA |
BamHI |
| SacI-gadB-F |
GGCgagctcATGTTTAAAGCTGTTCTGTTGGGCAA |
SacI |
| XhoI-gadBF-R | CCGctcgagTTACTTGTCATCGTCATCCTTGTAGTCAGGTCGGAACTACTCGATTCACG | XhoI |
Restriction enzyme cleavage sites are shown in small letters, and complementary sequences of the primer pairs used for overlap-extension PCR are shown in italics.
Construction of C. glutamicum pknG deletion mutants
C. glutamicum ATCC 13032 was transformed with pTM44-ΔpknG using a Gene Pulser Xcell electroporator (Bio-Rad, Richmond, CA, USA) (2.5 kV, 25 μF electric pulse in a 0.1-cm cuvette) followed by heat shock at 46°C for 6 min. The cells were then incubated in 1 mL of BHI medium at 30°C for 1.5 hours. After cultivation for 2 days at 30°C on BHI agar plates containing 25 μg mL−1 kanamycin, the transformants were selected for a strain with a single crossover of the ΔpknG genotype, which was then cultivated in 5 mL of BHI liquid medium at 30°C overnight and diluted 1:10,000 with MM medium (see below) containing 10% sucrose. The culture was plated on MM agar medium containing 10% sucrose and incubated for 2 days at 30°C. MM medium contains 1 g Yeast Extract, 10 g (NH4)2SO4, 1 g KH2PO4, 3 g urea, 0.4 g MgSO4∙7H2O, 2 mg FeSO4∙7H2O, 2 mg MnSO4∙5H2O, 0.05 g NaCl, 0.2 mg thiamine, and 0.05 mg biotin per liter. The occurrence of a double-crossover ΔpknG mutant was confirmed by its inability to grow after 1 day at 30°C on MM agar containing 25 μg mL−1 kanamycin. The sizes of ΔpknG (1.60-kbp) in the C. glutamicum genome was confirmed using directed PCR with the primer pairs used for the construction and KOD FX (Toyobo). The selected double-crossover strain was designated C. glutamicum ΔpknG.
Construction of C. glutamicum mutants that express GAD
The construction of the GAD-expression plasmid pCH-gadB, C. glutamicum GAD (strain ATCC 13032 harboring pCH-gadB), and C. glutamicum W (strain ATCC13032 harboring pCH) was reported (Takahashi et al. [2012]). Plasmid pCH is an E. coli-C. glutamicum shuttle vector that drives gene expression with a highly active constitutive promoter (Tateno et al. [2007]). The pCH-gadB construct was introduced into C. glutamicum ΔpknG. Transformants were selected by growth on BHI agar containing 25 μg mL−1 kanamycin. The presence of gadB was confirmed using directed PCR with the primer pair SacI-gadB-F and XhoI-gadBF-R. The resulting strain, C. glutamicum ΔpknG (pCH-gadB) was designated C. glutamicum GADΔpknG.
Western blotting analysis
C. glutamicum strains W, GAD, and GADΔpknG were cultured in test tubes at 30°C for 24 hours in 5 mL BHI medium containing 25 μg mL−1 kanamycin. Each culture (0.2 mL) was transferred to 20 mL BHI medium containing 25 μg mL−1 kanamycin in a 200 mL shaker flask. After fermentation for 24 hours, the cells from a 1 mL culture were centrifuged at 8,000 × g for 5 min, washed once in 50 mM Tris–HCl (pH 6.8) buffer, suspended in 1 mL of this buffer, and then 0.7 g of 0.1-mm diameter glass beads YGB01 (Yasui Kikai, Japan) was added to the tube. The cells were disrupted using a Shake Master Neo (Bio Medical Science) by shaking the tube three times at 1,500 rpm for 1 min at 1-min intervals. After centrifugation at 9,000 × g for 5 min, the supernatants were subjected to sodium dodecyl sulfate polyacrylamide gel electrophoresis analysis. The separated proteins were electroblotted onto a polyvinylidene fluoride membrane (Millipore, Boston, MA, USA) and then reacted sequentially with a mouse anti-FLAG M2 monoclonal antibody (Sigma, St. Louis, MO, USA) and a goat anti-mouse IgG alkaline phosphate conjugate (Promega) secondary antibody. The membrane was stained with 4-nitro-blue tetrazolium chloride (Promega) and 5-bromo-4-chloro-3-indolyl phosphate (Promega) according to the manufacturer’s instructions.
Culture conditions for GABA fermentation from glucose
To produce GABA, C. glutamicum GAD and the mutant strains were cultured in a test tube at 30°C for 22 hours in 5 mL BHI medium containing 25 μg mL−1 kanamycin. This culture (0.2 ml) was transferred to a 200 mL shaker flask containing 20 mL GABA Production 2 (GP2) medium with 25 μg mL−1 of kanamycin. GP2 medium contains 50 g glucose, 50 g (NH4)2SO4, 1 g K2HPO4, 3 g urea, 0.4 g MgSO4∙7H2O, 50 g soypeptone, 0.01 g FeSO4∙7H2O, 0.01 g MnSO4∙5H2O, 200 μg thiamine, 0.5 mg biotin, and 0.265 g pyridoxal 5′-phosphate (PLP) L−1. Stock solutions of thiamine, biotin, and PLP were filtered through a 0.22 μm membrane and added to the medium before adding cells. The initial pH of the GP2 medium was 6.30. The pH of the GP2 medium was not adjusted during the fermentation. The fermentation was performed in a BR-13FR BioShaker (Taitec, Japan) at 30°C at 120 rpm.
To determine the effect of adding glutamate (Figure 1), C. glutamicum GAD was cultured for 96 hours, and 1 g L−1 or 2 g L−1 of glutamate was added to the culture at 24 hours. To determine the effect of the pknG deletion on GABA production (Figure 2), C. glutamicum GAD and C. glutamicum GAD ΔpknG were cultivated for 96 hours. C. glutamicum W served as a control.
Figure 1.

Effect of glutamate on GABA production byC. glutamicumGAD. To examine the effect of adding L-glutamate during GABA fermentation, C. glutamicum GAD was cultured in 20 mL GP2 medium containing 25 μg mL−1 kanamycin in a shaker flask at 30°C for 96 hours. At 24 hours, 1.0 g L−1 (triangle) or 2.0 g L−1 (circle) of L-glutamate was added to the culture. GP2 medium without glutamate (diamond) served as a control. A: Extracellular GABA (solid line) and glucose (dotted line) levels in each flask were monitored for 96 hours. B: Extracellular glutamate production by C. glutamicum GAD in each flask was monitored throughout the fermentation. Data are expressed as the mean and standard error from three independent experiments.
Figure 2.

Time course of extracellular GABA production byC. glutamicumstrains GAD ∆pknG,C. glutamicumGAD, andC. glutamicumW. The three strains were cultured separately in 20 mL of GP2 medium containing 50 g L−1 glucose and 25 μg mL−1 kanamycin using a 200-mL shaker flask. Fermentation was performed at 30°C for 96 hours at 120 rpm. A: Extracellular GABA concentrations (circles) and glucose consumption (diamonds) of C. glutamicum strains GAD ∆pknG (closed symbols) and GAD (open symbols) were monitored. GABA (squares) and glucose consumption (dotted line) of strain W were monitored as controls. B: The OD600 values of C. glutamicum strains GAD ∆pknG (closed triangles) and GAD (open triangles) were monitored throughout the fermentation. Data are expressed as the mean and standard error from three independent experiments.
To determine the yield of GABA, fermentation was performed using strains GAD and GADΔpknG. The strains were cultivated in BHI medium at 30°C for 24 hours, and 5% (w/v) of the starter-culture solution was transferred to a 200 mL baffled flask containing 20 mL GP2 medium with 100 g L−1 glucose and 25 μg mL−1 kanamycin, and agitated at 120 rpm for 168 hours (Figure 3).
Figure 3.
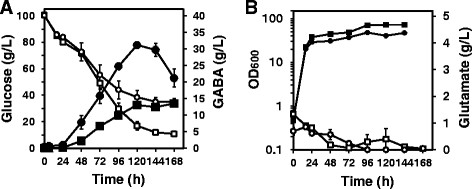
Time course of extracellular GABA production byC. glutamicumGAD, and GAD∆pknGfrom 100 g L−1glucose.C. glutamicum strains GAD, and GAD∆pknG were cultured separately in 20 mL of GP2 medium containing 100 g L−1 glucose and 25 μg mL−1 kanamycin in 200-mL baffled flasks. Fermentation was performed at 30°C for 168 hours at 120 rpm. A: Extracellular GABA concentrations (closed symbols) and glucose concentrations (open symbols) of C. glutamicum strains GAD (squares) and GAD∆pknG (circles ) were monitored. B: Extracellular glutamate concentrations (open symbols) and the OD600 (closed symbols) of the C. glutamicum strains GAD (squares), and GAD∆pknG (circles) were monitored throughout the fermentation. Data are expressed as the mean and standard error from three independent experiments.
Throughout cultivation, 1 ml of the culture was collected from each flask every 24 hours, centrifuged at 8,000 × g for 5 min at 4°C, and filtered through a 0.45 μm DISMIC Mixed Cellulose Ester (Advantec, Tokyo, Japan). The concentrations of GABA, glutamate, and glucose in culture supernatants were analyzed as described below. The optical density at 600 nm (OD600) was monitored simultaneously.
Analysis of cell growth, production of GABA and L-glutamate, and consumption of glucose
The growth of the C. glutamicum strains was monitored at OD600 using a UVmini-1240 UV–vis spectrophotometer (Shimadzu, Kyoto, Japan). GABA and L-glutamate concentrations in the supernatant were analyzed using a Shim-pack Amino-Li column (0.5 μm, 100 mm × 6.0 mm I.D, Shimadzu) and a Prominence Amino Acid Analyzer System (Shimadzu) after derivatization with ortho-phthalaldehyde. The mobile phase (lithium citrate-borate gradient ranging from pH 2.68–10.00) was delivered at 0.6 mL min−1 at 39°C. Amino acid mixtures Type AN-II and Type B (Wako Chemicals, Japan), 0.1 mM GABA (Nacalai Tesque), and 0.1 mM L-glutamate in sodium citrate buffer (pH 2.2) served as standards. Glucose concentrations were determined using a Prominence HPLC System (Shimadzu) equipped with an Shim-Pac SPR-Pb column (0.5 μm, 250 mm × 4.0 mm I.D., Shimadzu). Water served as the mobile phase and was delivered at a flow rate of 0.6 mL min−1 at 80°C. The elution profile was monitored using a refractometer.
Results
Effect of exogenous L-glutamate on GABA fermentation by C. glutamicum GAD
To examine the effect of glutamate as a precursor for GABA synthesis, L-glutamate was added to a culture of C. glutamicum GAD grown on GP2 medium with glucose as the primary carbon source, soy peptone as the nitrogen source, and the GAD cofactor PLP (Figure 1). The culture supernatant was periodically assayed for GABA, glucose, and glutamate (Figure 1). L-Glutamate was added to the culture of C. glutamicum GAD after 24 hours, and a parallel control culture lacked added L-glutamate (Figure 1B). The concentrations of GABA after 96 h of fermentation were 9.52 ± 1.14, 8.55 ± 0.2, and 7.49 ± 2.14 g L−1 in media containing either 2 g L−1, 1 g L−1 L-glutamate, or no glutamate, respectively. The maximum concentration of GABA produced by strain GAD in the presence of 2 g L−1 L-glutamate was 10.47 ± 0.41 g L−1 after 72 h (Figure 1A). The glutamate concentration in the medium of each culture decreased after 24 hours (Figure 1B), and the addition of glutamate was effective for prolonging GABA production.
Expression of GAD by pknG deletion mutants
C. glutamicum ΔpknG was constructed as described in the Materials and methods section. The GAD-expression plasmid pCH-gadB was introduced into C. glutamicum ΔpknG, and the resultant recombinant strain harboring pCH-gadB was named C. glutamicum GADΔpknG. C. glutamicum strains GAD and W served as controls. The intracellular expression levels of GAD in the engineered C. glutamicum strains were monitored by western blotting analysis using an antibody raised against the FLAG-tagged sequence that was incorporated into the cloned genes. The 53 kDa GadB band was detected in the cytoplasmic fractions prepared from C. glutamicum strains GAD, GADΔpknG, but not strain W (data not shown).
Influence of the pknG deletion on GABA synthesis
We next examined the effect of the pknG deletion on GABA production from glucose, using C. glutamicum strains GAD, GADΔpknG, and W. As glucose was consumed, GABA formation was detected in the media from cultures of each during stationary phase (Figure 2A). The GABA concentration in the supernatants of cultures of strain GADΔpknG reached 8.48 ± 0.30 g L−1 in 72 hours while strain GAD produced 5.79 ± 0.20 g L−1 of GABA (Figure 2A). The yield of GABA produced by strain GADΔpknG was 1.46-fold higher compared with that of strain GAD, suggesting that the pknG deletion reduced ODHC activity, causing an increase in GABA synthesis. Under these fermentation conditions, strains GADΔpknG and GAD consumed 44.92 g L−1 and 46.39 g L−1 of glucose within 72 hours, respectively (Figure 2A). The yields of GABA from glucose by strains GADΔpknG and GAD reached 0.337 mol mol−1 and 0.233 mol mol−1, respectively, in 72 hours. The growth rates of strain GADΔpknG were lower than that of strain GAD (Figure 2B). GABA formation was not observed in the culture medium of strain W (Figure 2A).
GABA fermentation by GAD and GAD∆pknG
To evaluate GABA production, strains GAD, and GAD∆pknG were separately cultivated in GP2 medium containing 100 g L−1 of glucose using baffled flasks (Figure 3). As glucose in the GP2 medium consistently decreased from the beginning of the fermentation, the concentration of extracellular GABA produced by C. glutamicum GAD∆pknG simultaneously increased, reaching a maximum level of 31.16 ± 0.41 g L−1 after 120 hours (Figure 3A). The rate of GABA production by C. glutamicum GAD∆pknG reached 0.259 (g L−1 h−1). As 60.90 ± 4.89 g L−1 of glucose was consumed by GAD∆pknG in 120 hours, the molar yield of GABA from glucose reached 0.893 mol mol−1 (Table 3). At the same time, strain GAD produced 13.06 ± 0.45 g L−1 of GABA in 120 hours, consuming 83.62 ± 2.92 g L−1 of glucose (Figure 3A, Table 3), The glucose consumption rate of strain GAD∆pknG was lower than that of strain GAD (Figure 3A). The molar yield of GABA from glucose by strain GAD was 0.272 mol mol−1 in 120 hours. Therefore, the yield of GABA produced by C. glutamicum GAD∆pknG increased 2.29-fold compared with that of C. glutamicum GAD (Table 3). The growth rate of strain GADΔpknG was lower than that of strain GAD. Extracellular L-glutamate was not produced by either strain GAD or GAD∆pknG using these fermentation conditions (Figure 3B).
Table 3.
Growth (OD600), GABA formation (gL−1, gL−1 h−1) and yield (mol GABA mol glucose−1) ofC. glutamicumstrains producing GABA for 120 hours
| C. glutamicum strain | GAD | GADΔ pknG |
|---|---|---|
| OD600 |
71.86 ± 0.59 |
40.56 ± 1.05 |
| Glucose consumed (gL−1) |
83.62 ± 2.92 |
60.90 ± 4.89 |
| GABA (gL−1) |
13.06 ± 0.45 |
31.16 ± 0.41 |
| Relative difference |
1 |
2.29 |
| GABA (gL−1 h−1) |
0.108 |
0.259 |
| Yield (mol GABA mol glucose−1) | 0.272 | 0.893 |
Discussion
In the present study, we established a robust system for producing GABA by deleting pknG from a strain of C. glutamicum that overexpresses GAD. In our previous study, GAD was introduced into wild-type C. glutamicum, because it overproduces the GABA precursor L-glutamate from sugar, whereas GABA is produced by C. glutamicum GAD directly from glucose. In our optimized conditions for GABA fermentation, GP2 medium contains biotin to support growth. The production of GABA suggests that intracellular glutamate is converted to GABA by strain GAD (Takahashi et al. [2012]). Wild-type C. glutamicum does not produce glutamate under ordinary culture conditions unless glutamate secretion is induced by culturing the biotin-auxotrophic wild-type strain in biotin-limiting conditions (Shiio et al. [1962]). Moreover, when L-glutamate was added to cultures of C. glutamicum GAD to determine its effect on GABA fermentation, an increase in the levels of GABA in the medium was observed when 2 g L−1 of L-glutamate was added (Figure 1A). We reasoned that because glutamate is a precursor in the synthesis of GABA, its increased availability would enhance the yield of GABA. Based on this rationale, we were able to successfully generate a C. glutamicum mutant that produced relatively high levels of GABA.
We focused on pknG, because its product (PknG) regulates the activity of ODHC. PknG activates ODHC by phosphorylating its subunit OdhI, which is a subunit of ODHC. ODHC acts at a branch point of the TCA cycle where it catalyzes the conversion of 2-oxoglutarate to succinyl-CoA. Unphosphorylated OdhI inhibits the ODHC activity of C. glutamicum (Niebisch et al. [2006]). A pknG-deficient mutant of C. glutamicum produces 4.3-fold higher amounts of glutamate compared with wild-type under biotin-limiting conditions (Schultz et al. [2007]). The C. glutamicum 2262 pknG mutant also produces glutamate at a 40% higher specific rate compared with wild-type (Boulahya et al. [2010]). Therefore, a pknG-deficient strain was constructed to reduce the metabolic flux to the TCA cycle.
We show here that the yield of GABA in cultures of strain GAD∆pknG was 2.29-fold higher in 120 hours compared with that of strain GAD (Figure 3, Table 3), suggesting that the pknG deletion influenced ODHC activity by causing an increase in the intracellular glutamate level that enhanced GABA production. We assumed that the ODHC activity of strain GAD∆pknG was reduced, because OdhI was not phosphorylated and could not activate the ODHC complex, which caused an increase in carbon flux into the glutamate pathway compared with that of strain GAD. The glucose consumption rate and growth rate of GAD∆pknG was lower than that of GAD (Figure 3, Table 3), suggesting that the flux to TCA cycle was decreased. We plan to analyze carbon flux of these strains in the future. Moreover, in the late stage of fermentation, reduction of GABA production was observed in cultures of strain GAD∆pknG (Figure 3A). Because reduced levels of the product were also observed in cultures of strain GAD (Takahashi et al. [2012]), we are now attempting to disrupt the genes for GABA assimilation.
In our GABA production system using C. glutamicum GADΔpknG, high concentrations of GABA were produced from glucose in GP2 medium without the addition of glutamate. The yield of GABA from glucose produced by strain GADΔpknG reached 0.893 mol mol−1, and the highest yield was produced in 120 hours (Table 3). Using C. glutamicum GADΔpknG, we expect that fewer fermentation by-products will be produced and that the recovery of GABA will be simpler than using methods for its isolation from cultures of wild-type lactic acid bacteria.
GABA is primarily produced using cultures of lactic acid bacteria containing glutamate or monosodium glutamate (MSG) (Li and Cao [2010]). For example, Streptococcus salivarius subsp. thermophilus Y2, a cheese starter strain, produces 7.98 g L−1 of GABA after 84 hours of fermentation with a continuous supply of 15 g L−1 MSG, corresponding to a rate of 0.095 g L−1 hour−1 (Lu et al. [2008]). L. paracasei NFRI 7415, which was isolated from fermented fish, produces 31.11 g L−1 (302 mM) GABA in 168 hours, corresponding with a production rate of 0.185 g L−1 hour−1. Although the production rate by strain NFRI 7415 was relatively high, 500 mM (73.5 g L−1) glutamate was added to the culture medium (Shima et al. [2005]). L. brevis NCL912, which was isolated from Paocai, produces 345.83 mM (35.6 g L−1) GABA in a medium containing 500 mM glutamate (Cao et al. [2010]). Microbial production systems that require supplementation with amino acids are not cost-effective for applications such as synthesizing chemicals. In contrast, in our GABA production system using C. glutamicum GADΔpknG, high concentrations of GABA were produced from glucose in one step (31.16 g L−1 from glucose at 0.259 g L−1 h−1 and therefore will provide a new platform for synthesizing chemicals.
Recently, two GADs from L. brevis were expressed in C. glutamicum ATCC 13032/pDXW-8-gadRBC2, which produced 27.13 ± 0.54 g L−1 of GABA from glucose in 120 hours using flask fermentation with six urea supplements to the medium during the fermentation (Shi et al. [2013]). With urea supplementation, increased amounts of glutamate were produced in the culture supernatant at the same time; however, it may be difficult to separate the product from the medium. In our system, 31.16 g L−1 of GABA was directly produced from glucose without addition of a nitrogen or carbon source during the fermentation. Notably, because our GP2 medium contains biotin to support growth, glutamate is not secreted (Figures 3B and 4). A one-step production system has long been a goal for producing precursors for synthesizing bulk chemicals, and we show here that this was possible for robust production of GABA using GADΔpknG.
Figure 4.
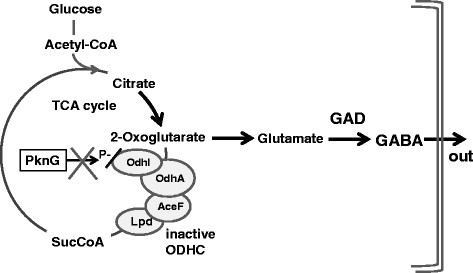
Model of direct GABA production by C. glutamicum GAD∆ pknG .
Our future work involves the development of a process to produce GABA from abundantly available starch or cellulose. We developed a system for coexpressing amylase and lysine decarboxylase in C. glutamicum to produce cadaverine from soluble starch (Tateno et al. [2009]). Further, our C. glutamicum endoglucanase secretion systems for producing glutamate from beta glucan (Tsuchidate et al. [2011]) can be applied to the production of GABA. The production of GABA using strains based on C. glutamicum GADΔpknG would allow the synthesis of 100% biomass-derived nylon PA4. Notably, C. glutamicum is generally recognized as safe (GRAS) according the United States Food and Drug Administration. Therefore, the system for GABA fermentation developed in the present study can be applied to the production of GABA as a component of foods and pharmaceuticals.
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Naoko Okai, Email: okai@port.kobe-u.ac.jp.
Chihiro Takahashi, Email: 111t435t@stu.kobe-u.ac.jp.
Kazuki Hatada, Email: 094t457t@stu.kobe-u.ac.jp.
Chiaki Ogino, Email: ochiaki@port.kobe-u.ac.jp.
Akihiko Kondo, Email: akondo@kobe-u.ac.jp.
Acknowledgment
This work was partially supported by the Special Coordination Funds for Promoting Science and Technology, Creation of Innovation Centers for Advanced Interdisciplinary Research Areas (Innovative Bioproduction Kobe) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- Boulahya KA, Guedon E, Delaunay S, Schultz C, Boudrant J, Bott M, Goergen JL. OdhI dephosphorylation kinetics during different glutamate production processes involving Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2010;4:1867–1874. doi: 10.1007/s00253-010-2599-y. [DOI] [PubMed] [Google Scholar]
- Cao YS, Li HX, Qiu T, Gao DD. Medium optimization for production of gamma-aminobutyric acid by Lactobacillus brevis NCL912. Amino Acids. 2010;4:1439–1445. doi: 10.1007/s00726-009-0355-3. [DOI] [PubMed] [Google Scholar]
- DeBiase D, Tramonti A, John RA, Bossa F. Isolation, overexpression, and biochemical characterization of the two isoforms of glutamic acid decarboxylase from Escherichia coli. Protein Expr Purif. 1996;4:430–438. doi: 10.1006/prep.1996.0121. [DOI] [PubMed] [Google Scholar]
- Fonda ML. L-glutamate decarboxylase from bacteria. Methods Enzymol. 1985;4:11–16. doi: 10.1016/S0076-6879(85)13005-3. [DOI] [PubMed] [Google Scholar]
- George M. Burgey’s Manual of Systematic Bacteriology. Springer, New York; 2001. [Google Scholar]
- Hashimoto K, Hamano T, Okada M. Degradation of several polyamides in soils. J Appl Polym Sci. 1994;4:1579–1583. doi: 10.1002/app.1994.070541023. [DOI] [Google Scholar]
- Hayakawa K, Kimura M, Kasaha K, Matsumoto K, Sansawa H, Yamori Y. Effect of a gamma-aminobutyric acid-enriched dairy product on the blood pressure of spontaneously hypertensive and normotensive Wistar-Kyoto rats. Br J Nutr. 2004;4:411–417. doi: 10.1079/BJN20041221. [DOI] [PubMed] [Google Scholar]
- Hermann T. Industrial production of amino acids by Coryneform bacteria. J Biotechnol. 2003;4:155–172. doi: 10.1016/S0168-1656(03)00149-4. [DOI] [PubMed] [Google Scholar]
- Kawasaki N, Nakayama A, Yamano N, Takeda S, Kawata Y, Yamamoto N, Aiba S. Synthesis, thermal and mechanical properties and biodegradation of branched polyamide 4. Polymer. 2005;4:9987–9993. doi: 10.1016/j.polymer.2005.06.092. [DOI] [Google Scholar]
- Kimura E. Triggering mechanism of L-glutamate overproduction by dtsR1 in Coryneform bacteria. J Biosci Bioeng. 2002;4:545–551. doi: 10.1016/s1389-1723(02)80193-1. [DOI] [PubMed] [Google Scholar]
- Leuchtenberger W, Huthmacher K, Drauz K. Biotechnological production of amino acids and derivatives: Current status and prospects. Appl Microbiol Biotechnol. 2005;4:1–8. doi: 10.1007/s00253-005-0155-y. [DOI] [PubMed] [Google Scholar]
- Li HX, Cao YS. Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids. 2010;4:1107–1116. doi: 10.1007/s00726-010-0582-7. [DOI] [PubMed] [Google Scholar]
- Lu ZX, Yang SY, Lu FX, Bie XM, Jiao Y, Sun LJ, Yu B. Production of gamma-aminobutyric acid by Streptococcus salivarius subsp. thermophilus Y2 under submerged fermentation. Amino Acids. 2008;4:473–478. doi: 10.1007/s00726-007-0544-x. [DOI] [PubMed] [Google Scholar]
- Mimitsuka T, Sawai H, Hatsu M, Yamada K. Metabolic engineering of Corynebacterium glutamicum for cadaverine fermentation. Biosci Biotechnol Biochem. 2007;4:2130–2135. doi: 10.1271/bbb.60699. [DOI] [PubMed] [Google Scholar]
- Niebisch A, Kabus A, Schultz C, Weil B, Bott M. Corynebacterial protein kinase G controls 2-oxoglutarate dehydrogenase activity via the phosphorylation status of the OdhI protein. J Biol Chem. 2006;4:12300–12307. doi: 10.1074/jbc.M512515200. [DOI] [PubMed] [Google Scholar]
- Oda KH, Hiraga K, Ueno YH. Glutamate decarboxylase from Lactobacillus brevis: Activation by ammonium sulfate. Biosci Biotechnol Biochem. 2008;4:1299–1306. doi: 10.1271/bbb.70729. [DOI] [PubMed] [Google Scholar]
- Schultz C, Niebisch A, Gebel L, Bott M. Glutamate production by Corynebacterium glutamicum: Dependence on the oxoglutarate dehydrogenase inhibitor protein OdhI and protein kinase PknG. Appl Microbiol Biotechnol. 2007;4:691–700. doi: 10.1007/s00253-007-0933-9. [DOI] [PubMed] [Google Scholar]
- Schultz C, Niebisch A, Schwaiger A, Viets U, Metzger S, Bramkamp M, Bott M. Genetic and biochemical analysis of the serine/threonine protein kinases PknA, PknB, PknG and PknL of Corynebacterium glutamicum: Evidence for non-essentiality and for phosphorylation of OdhI and FtsZ by multiple kinases. Mol Microbiol. 2009;4:724–741. doi: 10.1111/j.1365-2958.2009.06897.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi F, Jiang J, Li Y, Xie Y. Enhancement of gamma-aminobutyric acid production in recombinant Corynebacterium glutamicum by co-expressing two glutamate decarboxylase genes from Lactobacillus brevis. J Ind Microbiol Biotechnol. 2013;4:1285–1296. doi: 10.1007/s10295-013-1316-0. [DOI] [PubMed] [Google Scholar]
- Shiio I, Otsuka SI, Takahashi M. Effect of biotin on the bacterial formation of glutamic acid I. Glutamate formation and cellular permeability of amino acids. J Biochem. 1962;4:56–62. doi: 10.1093/oxfordjournals.jbchem.a127500. [DOI] [PubMed] [Google Scholar]
- Shima J, Komatsuzaki N, Kawamoto S, Momose H, Kimura T. Production of gamma-aminobutyric acid (GABA) by Lactobacillus paracasei isolated from traditional fermented foods. Food Microbiol. 2005;4:497–504. doi: 10.1016/j.fm.2005.01.002. [DOI] [Google Scholar]
- Shima J, Komatsuzaki N, Nakamura T, Kimura T. Characterization of glutamate decarboxylase from a high gamma-aminobutyric acid (GABA)-producer, Lactobacillus paracasei. Biosci Biotechnol Biochem. 2008;4:278–285. doi: 10.1271/bbb.70163. [DOI] [PubMed] [Google Scholar]
- Takahashi C, Shirakawa J, Tsuchidate T, Okai N, Hatada K, Nakayama H, Tateno T, Ogino C, Kondo A. Robust production of gamma-amino butyric acid using recombinant Corynebacterium glutamicum expressing glutamate decarboxylase from Escherichia coli. Enzyme Microb Technol. 2012;4:171–176. doi: 10.1016/j.enzmictec.2012.05.010. [DOI] [PubMed] [Google Scholar]
- Tateno T, Fukuda H, Kondo A. Direct production of L-lysine from raw corn starch by Corynebacterium glutamicum secreting Streptococcus bovis alpha-amylase using cspB promoter and signal sequence. Appl Microbiol Biotechnol. 2007;4:533–541. doi: 10.1007/s00253-007-1191-6. [DOI] [PubMed] [Google Scholar]
- Tateno T, Okada Y, Tsuchidate T, Tanaka T, Fukuda H, Kondo A. Direct production of cadaverine from soluble starch using Corynebacterium glutamicum coexpressing alpha-amylase and lysine decarboxylase. Appl Microbiol Biotechnol. 2009;4:115–121. doi: 10.1007/s00253-008-1751-4. [DOI] [PubMed] [Google Scholar]
- Tsuchidate T, Tateno T, Okai N, Tanaka T, Ogino C, Kondo A. Glutamate production from beta-glucan using endoglucanase-secreting Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2011;4:895–901. doi: 10.1007/s00253-011-3116-7. [DOI] [PubMed] [Google Scholar]
- Yamano N, Nakayama A, Kawasaki N, Yamamoto N, Aiba S. Mechanism and characterization of polyamide 4 degradation by Pseudomonas sp. J Polym Environ. 2008;4:141–146. doi: 10.1007/s10924-008-0090-y. [DOI] [Google Scholar]


