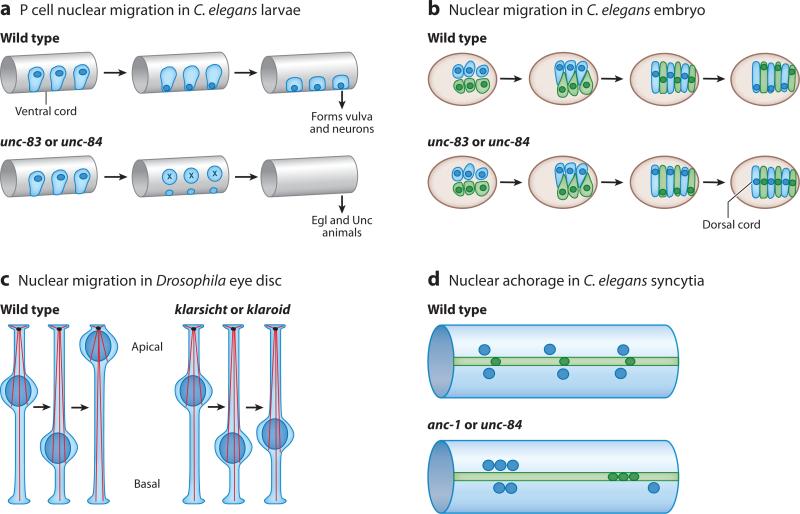
Figure 2.
Genetic models for studying nuclear positioning. (a) Nuclear migration in hypodermal P cells during the first larval stage of Caenorhabditis elegans. Nuclei (dark blue) migrate from a lateral to a ventral position through the cytoplasm (light blue). P cells normally go on to form the vulva and neurons in the ventral cord. Mutations in unc-83 or unc-84 disrupt nuclear migration and the P cells die, resulting in egg-laying defective (Egl) and uncoordinated (Unc) animals. (b) Nuclear migration in C. elegans embryonic hypodermal cells. Right (light blue) and left (light green) hyp7 precursors align along the dorsal cord of a pre-elongation embryo (brown circle, dorsal view, anterior to the left) and intercalate to form a row of column-shaped cells spanning the dorsal midline. Nuclei (dark blue and dark green) then migrate the length of the cell from right to left (blue) or left to right (green). Mutations in unc-83 and unc-84 disrupt nuclear migration, and all nuclei end up in the dorsal cord instead of their normal, lateral positions. (c) Nuclear migration in the Drosophila eye disc. After the morphogenetic furrow passes, nuclei (dark blue) migrate from a basal position to an apical one as they develop into photoreceptors. In klarsicht or klaroid mutants, nuclei abnormally remain basal, disrupting the development of the eye, whereas centrosomes (black ovals) organize microtubules (red strands) from an apical position. (d ) Nuclear anchorage in the adult C. elegans syncytial hypodermis. Three large hypodermal syncytia, the hyp7 (light blue) syncytium and the lateral seam-cell syncytia (light green), are used to study nuclear anchorage. Normally nuclei (dark blue and dark green) are anchored and spaced evenly apart. In anc-1 or unc-84 mutant animals, nuclei are unanchored and often associate in clusters.