Figure 3.
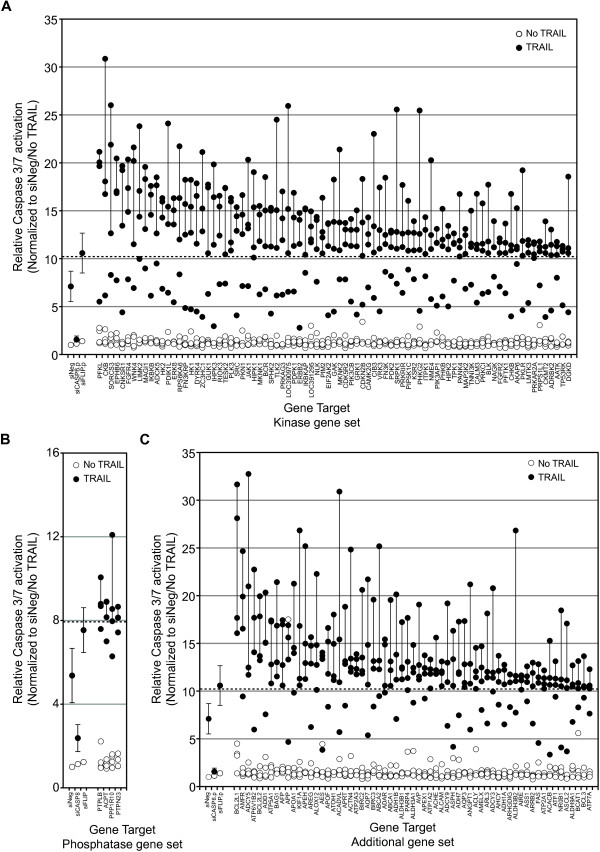
The identification of putative negative regulators of TRAIL-induced apoptosis by using siRNA-enhanced activation of caspase-3/7 in the presence of TRAIL. Genes for which three or four siRNAs mediated a fold change in the activation of caspase-3/7 in the presence of TRAIL of two or more standard deviations more than that observed in control siRNA (siNeg)-transfected cells plus TRAIL were considered putative negative regulators of TRAIL-induced apoptosis. The dashed line in each panel indicates the 2- standard deviation fold change, and a vertical line joins those siRNAs that induced at least this level of change for each gene. Data for the control siRNAs, siNeg, siCASP8, and siFLIP are included for reference and represent the same data for the siRNAs, indicated as shown in Figure 2Aii and Additional file 3: Figure S1Ai. (A) Genes identified from the kinase gene set. (B) Genes identified from the phosphatase gene set, and (C) genes identified from the additional gene set. Genes are ranked in descending order based on the median value for each set of four siRNAs per gene.
