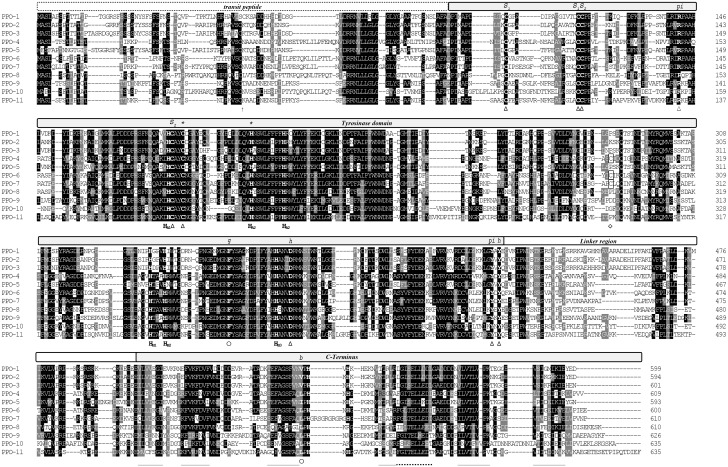
Figure 4. Sequence analysis of the dandelion PPO family.
Amino acid sequences of all PPOs were aligned using MUSCLE v3.7 [28]. Identical and similar residues are shaded in black and gray, respectively. Domain structure was analyzed [13], [43]–[44] and findings were marked as follows. The predicted transit peptide is labelled and the predicted cleavage site of the stromal processing peptidase is marked with an arrow. The catalytic tyrosinase domain, linker region, C-terminal domain, and copper-binding histidines of the CuA (HA1–HA3) and CuB (HB1–HB3) sites are labelled. Structurally important residues are marked by triangles (Δ) and potentially regulative residues by circles (○). The predicted β-strands of the conserved β-sandwich C-terminal domain are marked by straight underlining ( _ ) and the conserved helix by dotted underlining (…). Cysteines of potential multimerization sites [20] are boxed and their position is marked by a diamond (◊). Cysteines potentially involved in two intramolecular disulfide bonds are labeled S1 and S2. [S, disulfide linkage; *, thioether bridge; h, hydrogen bond; pi, π-cation interaction; g, gate residue; b, blocking residue].