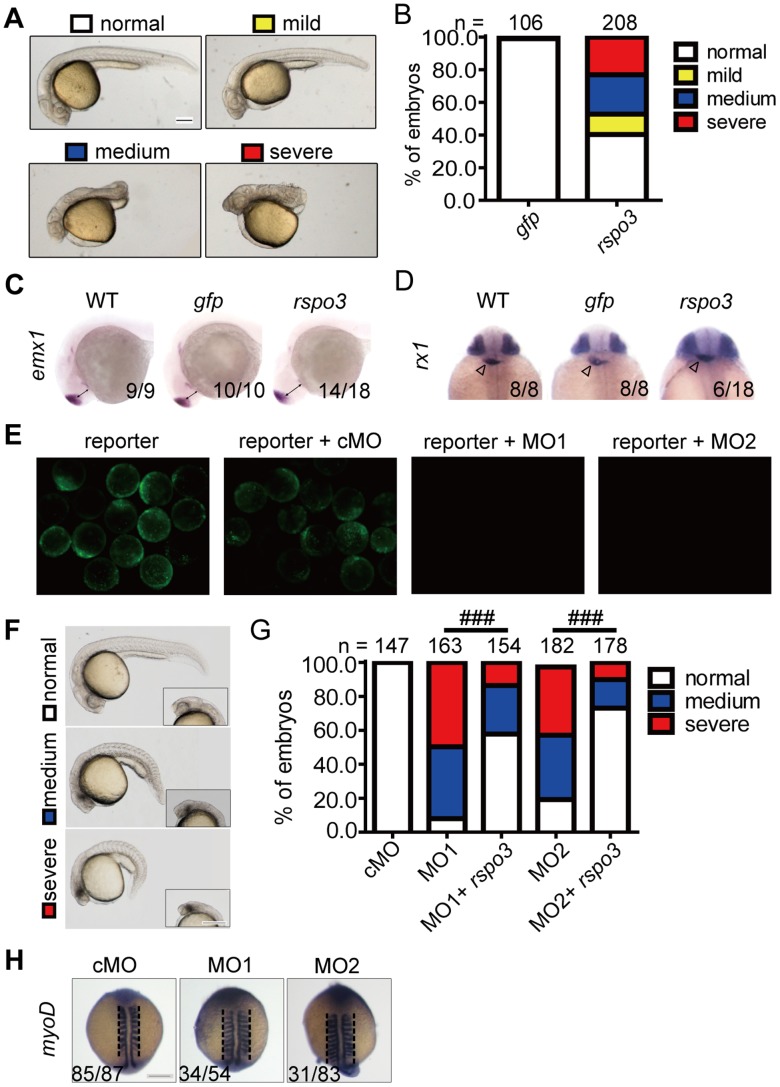
Figure 3. Effects of rspo3 overexpression and knockdown in zebrafish embryos.
(A) Classification of phenotypes caused by forced expression of rspo3. One-cell stage embryos were injected with 600 pg rspo3 mRNA. Embryos were raised to 24 hpf and examined. Lateral views with anterior to the left. Scale bar = 200 µm. (B) The percentages of embryos in each category as shown in (A). The results are from three independent experiments and the total embryo numbers are given at the top. (C, D) Expression patterns of the indicated marker genes in wild-type (WT) embryos or embryos injected with 600 pg gfp mRNA or rspo3 mRNA. Embryos were analyzed at 24 hpf by in situ hybridization. Lateral view with anterior to the left (C) and dorsal view with anterior up (D) are shown, and the frequency of embryos with the indicated patterns is shown in the bottom right in each panel. Double arrow lines in C show the distance from the telencephalon to the yolk. Blank arrow heads in D indicate the heart labeled by nkx2.5 mRNA. (E) Effectiveness and specificity of MOs used. Fluorescent micrographs of zebrafish embryos at 12 hpf injected with the rspo3 5′-UTR reporter plasmid alone (100 pg), the reporter plasmid DNA with control MO (4 ng), rspo3 targeting MO1 (4 ng) or rspo3 targeting MO2 (8 ng), respectively. (F) Classifications of phenotypes caused by morpholino-mediated knockdown of rspo3. Representative views of zebrafish embryos at 24 hpf injected with 8 ng control MO (cMO), 4 ng (MO1) or 8 ng (MO2) rspo3 targeting MO, and 4 ng MO1 or 8 ng MO2 plus 20 pg rspo3 mRNA (MO+rspo3). Lateral views with anterior to the left. The amplified head region of each embryo is shown in right corner insert. Scale bar = 200 µm. (G) The percentages of embryos in each category as shown in (F). The results are from three independent experiments and the total embryo numbers are given at the top. ### P<0.0001, Chi-Square test. (H) Expression patterns of the indicated marker genes in embryos injected with 8 ng cMO, 4 ng rspo3 MO1, or 8 ng MO2. Embryos were analyzed at 14 hpf by in situ hybridization. Dorsal view with anterior to the top is shown, and the frequency of embryos with the indicated patterns is shown in the bottom left corner of each panel. The blank dash lines show the extension of the marker expression. Scale bar = 200 µm. Next, knockdown experiments were carried out using two independent translation- blocking antisense MOs. The efficacy of these rspo3 targeting MOs was verified by co-injecting an rspo3 5′-UTR-GFP expression construct. Both MO1 and MO2 blocked the reporter GFP expression (Fig. 3E). Knockdown of rspo3 by either MO1 or MO2 resulted in an increase in the number of embryos displaying enhanced ventral-posterior phenotypes (Fig. 3F and 3G). In addition, knockdown of rspo3 resulted in lateral expansion of somites as indicated by myoD mRNA expression at 14 hpf (Fig. 3H). The abnormal embryos were classified into medium and severe groups (Fig. 3F and 3G). Embryos in the medium group exhibited smaller eyes, slightly reduced head, and curved body axis (Fig. 3F). Embryos in the severe group exhibited smaller eyes, reduced brain, and shorter and curved body axis (Fig. 3F). 50% and 42% of the MO1-injected embryos were in the severe and medium group (Fig. 3G). Likewise, 40% and 38% of the MO2-injected embryos were in the severe and medium group (Fig. 3G). Importantly, co-injection of rspo3 mRNA with MO1 or MO2 markedly reduced the MO-induced abnormal phenotypes from ∼90% to ∼40% (MO1) and ∼80% to ∼25% (MO2), respectively (p<0.0001) (Fig. 3G).