Figure 3.
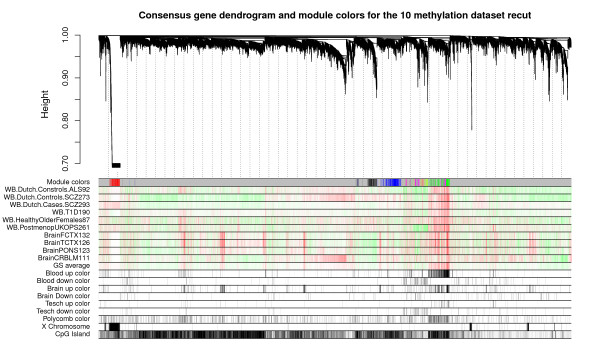
Hierarchical cluster tree and consensus module structure. Hierarchical cluster tree (dendrogram) of the consensus network based on ten independent methylation data sets. The first color band underneath the tree indicates the module color of each CpG site. The color grey is reserved for 'background' CpG sites that are not clustered into any module. The remaining color bands represent each gene's correlation with age in the underlying data sets; high intensity red values represent a strong positive correlation whereas high intensity green values represent a strong negative correlation. The remaining color bands indicate whether a gene was part of the core aging signature from Teschendorff et al. [16]. The color bands 'Tesch up' and 'Tesch down' indicate that Teschendorff et al. determined that methylation levels of this CpG site correlated positively or negatively with age, respectively. Other color bands indicate whether the CpG site is close to a known polycomb group target, is located on the X chromosome, or located in a CpG island. The figure suggests that the green module is composed of CpG sites that positively correlate with age in all ten tissues, which is why we refer to it as an aging module. Further, this aging related module is enriched with CpG sites that are close to Polycomb group target genes. Also note the presence of a very distinct red module that corresponds to CpG sites located on the X chromosome.
