Figure 6.
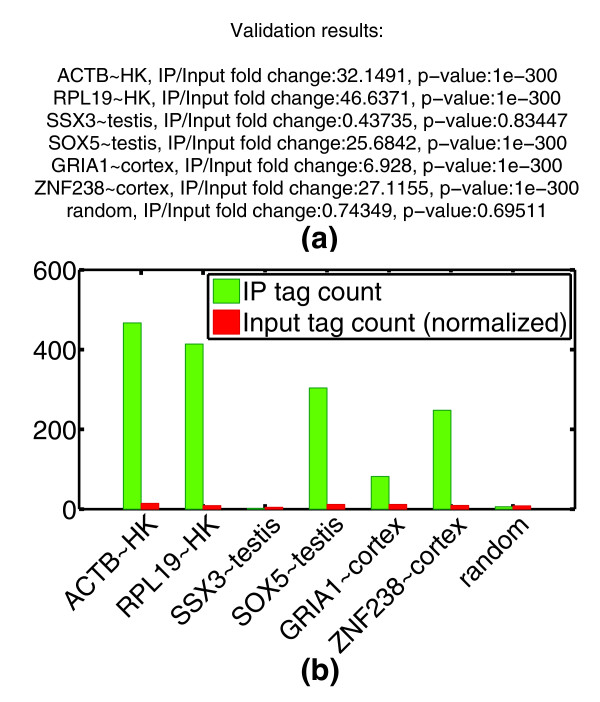
Spot validation. This figure demonstrates CHANCE's spot validation module. H3K4me3 in H1 HESCs from the Broad ENCODE data is spot validated for promoter regions of known housekeeping and tissue-specific genes. (a) The summary statement gives the IP over Input fold-change in read count as well as a P-value based on a Poisson null model (see Materials and methods). A random locus is added for comparison. The putative proximal promoter was estimated to 3 kbp upstream to 3 kbp downstream of the transcription start site. (b) Graphical representation of the results. On the x-axis, we have the gene symbols followed by the tissue type with which their expression is commonly identified; HK denotes 'house keeping' or ubiquitously expressed genes. The y-axis shows the number of reads mapping to the corresponding promoter region, both in IP and Input.
