Figure 4.
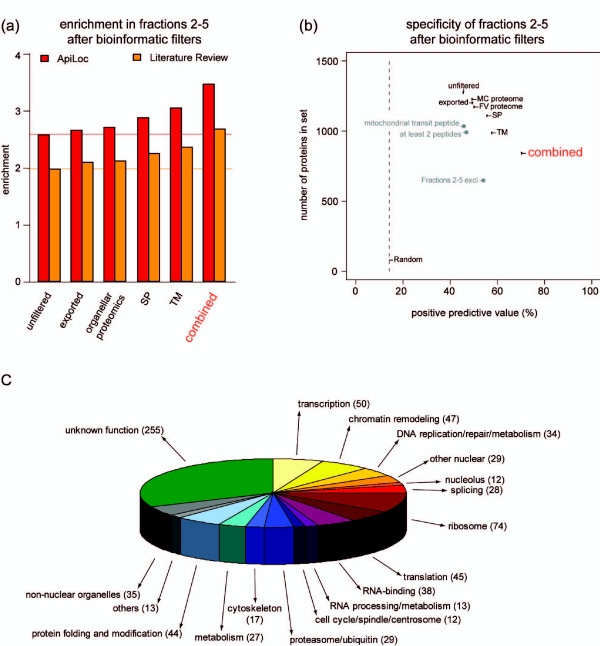
Bioinformatic filtering of the nuclear proteomics data. (a) Postprocessing enrichments in the set of proteins found in fractions 2 to 5. (b) Nuclear protein content in fractions 2 to 5 after bioinformatic filtering. exported, removal of proteins predicted to be exported; organellar proteomics, removal of proteins detected in the Maurer's cleft (MC) [53] and/or the food vacuole (FV) proteome [52]; SP, removal of proteins carrying a predicted SP; TM, removal of proteins carrying predicted TM domains; combined, the five accepted filters (SP, TM, FV, MC, EXP) combined; mitochondrial transit peptides, removal of proteins predicted by PlasMit [67]; at least two peptides, removal of proteins detected by a single peptide only; fractions 2-5 excl., removal of proteins detected in the cytoplasmic fraction. (c) Classification of the core nuclear proteome into broad generic functional classes was done by manual inspection of the dataset and is based on functional annotation available on PlasmoDB [68] and literature review. Each protein was assigned to a single class only.
