Figure 5.
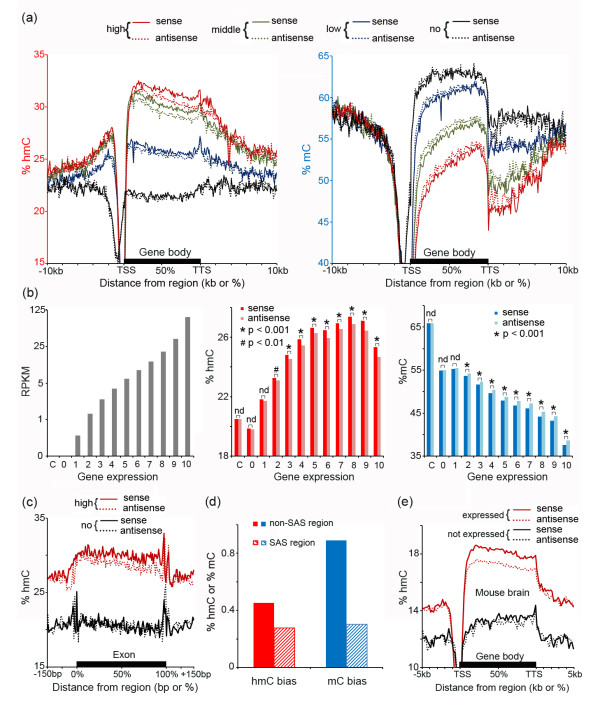
Strand-biased hmC and mC profiles on the gene body. (a) Profiles of hmC (left panel) and mC (right panel) on the sense (lined) and antisense (dotted) strands of the genes expressed at high (red), middle (green) and low (blue) expression levels, as well as the genes with no expression (black). TSS and TTS indicate the transcription starting site and the transcription terminal site, respectively. (b) Expressed genes were divided into 10 groups according to the expression levels (left panel), and the average levels of hmC (middle panel) and mC (right panel) for each strand of the gene body were measured. The values for the genes that are not expressed (expression level 0) and randomly selected intergenic regions as the control (C) are also shown. One-tailed paired Student’s t test. nd, no statistical difference (P >0.05). (c) Profiles of hmC on the sense (lined) and antisense (dotted) strands of exons with high (red) or no (black) expression. (d) The hmC and mC strand-biases are reduced at the sense-antisense gene (SAS) paired regions in comparison with the non-SAS regions. (e) The hmC profile on the sense (lined) and antisense (dotted) strands of the genes that are expressed (red) or not expressed (black) in the mouse brain exhibits the transcription-correlated hmC bias toward the sense strand similar to the human pattern. The TAB-Seq, BS-Seq, and RNA-Seq data for analysis were obtained from Lister et al.[25].
