Figure 5.
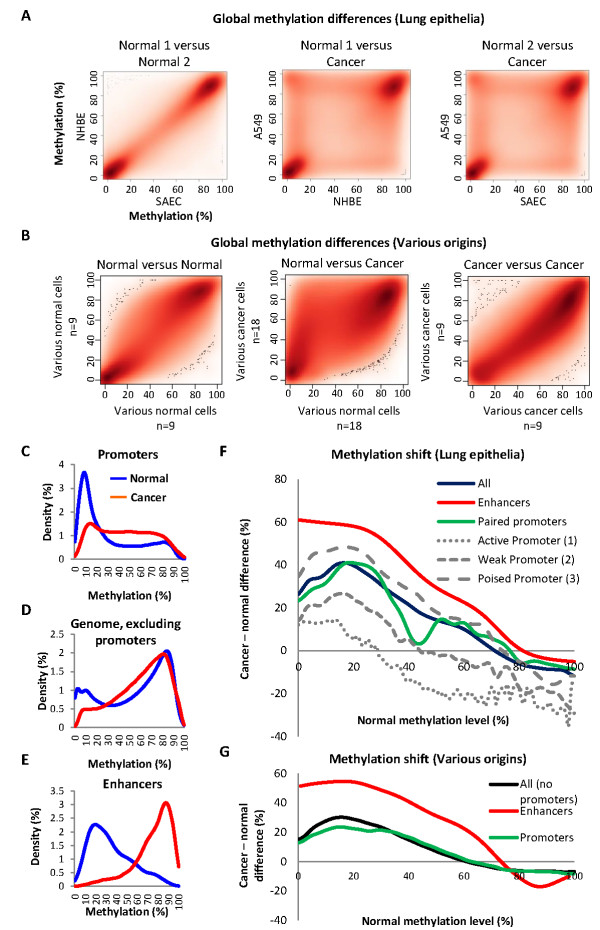
Enhancer methylation maybe specifically altered in cancer. (A,B)x-y scatters showing the methylation of sites across the genome in a given cell type (or a collection of cell types) versus another cell type (or types).(A)Normal and cancerous lung epithelia: genome-wide methylation of normal versus normal (left) or of cancer versus normal (middle and right) cell types. The given cell types are indicated, as listed in table S2. (B) Normal and cancerous cell types of various tissue origins: genome-wide methylation of nine normal cell types of various tissue origins, versus another nine cell types (left), of 18 cancer versus 18 normal cell types (middle), or of nine cancers versus other cancers (right). (C-E) Distributions of methylation levels in the collection of normal and cancer samples shown in B, in promoter sites, in all sites excluding promoters, or in the high-scoring enhancer sites. (F) Average gain or loss of methylation in cancer, as a function of the methylation levelin normal cells: for any given level of methylation in the normal cells, the graph shows the average change in the cancer, according to the global trend shown in B. Data are shown for all sites; for sites in the high-scoring enhancers; for sites in the promoters paired with the high-scoring enhancers; or for sites in strong, weak or poised promoters. The analysis is based on methylation levels and chromatin states in human mammary epithelial cells and MCF7 cells shown in A. (G)Same analysis as in F, but for 18 normal and 18 cancer cell types shown in B.
