Figure 3.
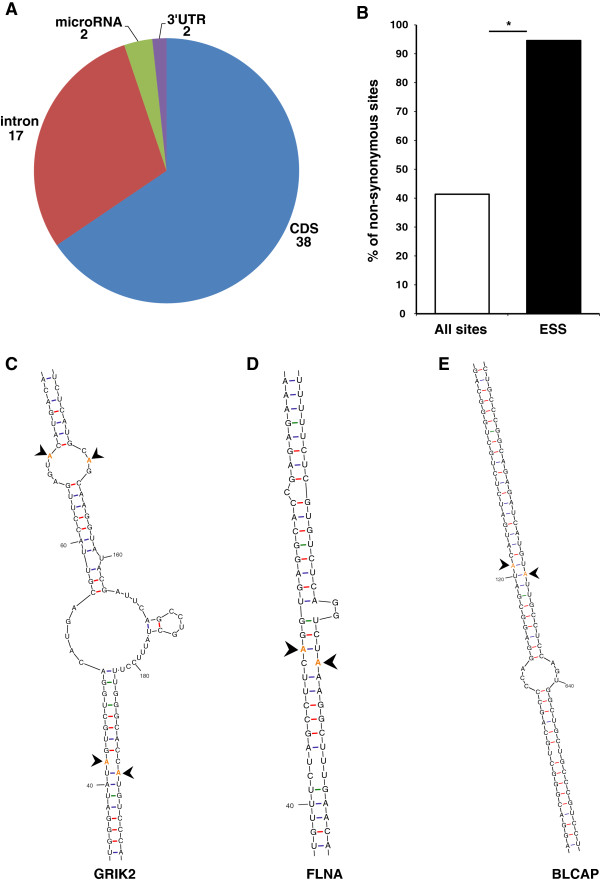
Most of the ESS sites are located in a coding region or adjacent to such a site. (A) Genomic location of evolutionarily conserved sites. (B) Frequency of non-synonymous editing alterations in exonic sites for both groups demonstrates enrichment of sites that cause amino acid change in the ESS compared to the control (all other sites, P <2 × 10-11 calculated by Fisher’s exact test). (C-E) Secondary structure shows spatial proximity of coding and intron sites of GRIK2 (C), FLNA (D) and BLCAP (E) genes; editing sites are highlighted in orange and marked by an arrow.
