Figure 2.
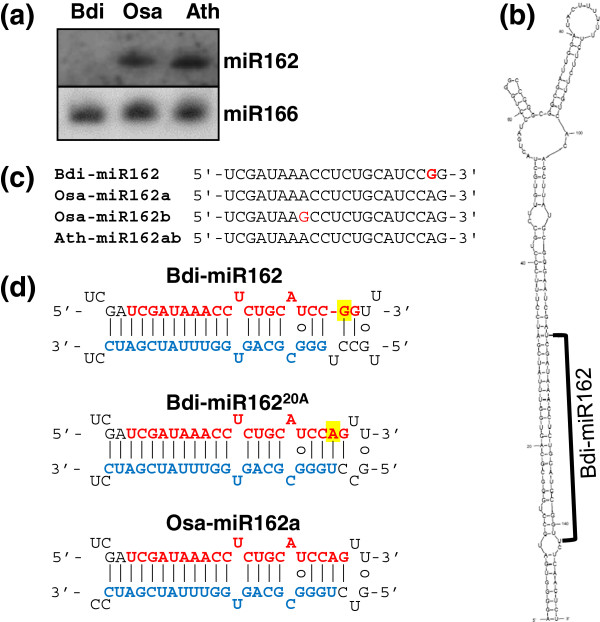
Characteristics of Bdi-miR162. (a) RNA blot of miR162 for Brachypodium (Bdi), rice (Osa) and Arabidopsis (Ath). Mixed probes of P32-end labeled Bdi-miR162 and Osa-miR162a were used to detect mature miR162 from Brachypodium panicle, rice panicle and Arabidopsis flower tissues. miR166 was used as a control. (b) Predicted secondary structure of the Bdi-MIR162 precursor. The location of Bdi-miR162 is indicated with a black line. (c) Sequence alignment of miR162 for Brachypodium, rice and Arabidopsis. Nucleotides that differ are shown in red. (d) Comparison of base pairings in the folded MIR162 precursor of Brachypodium and rice. Base pairing in a mutated Bdi-MIR162 precursor is shown between them. Position 20 of Bdi-miR162 is highlighted in yellow. miRNA is shown in red and miRNA* in blue. miRNA, microRNA.
