Figure 1.
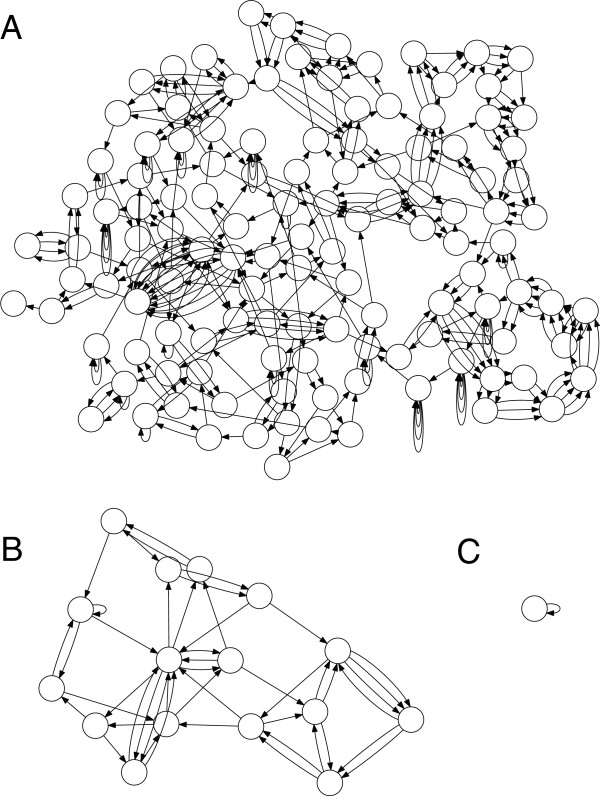
Genome assembly graph complexity is reduced as sequence length increases. Three de Bruijn graphs for E. coli K12 are shown for k of 50, 1,000, and 5,000. The graphs are constructed from the reference and are error-free following the methodology of Kingsford et al.[27]. Non-branching paths have been collapsed, so each node can be thought of as a contig with edges indicating adjacency relationships that cannot be resolved, leaving a repeat-induced gap in the assembly. (A) At k = 50, the graph is tangled with hundreds of contigs. (B) Increasing the k-mer size to k = 1,000 significantly simplifies the graph, but unresolved repeats remain. (C) At k = 5,000, the graph is fully resolved into a single contig. The single contig is self-adjacent, reflecting the circular chromosome of the bacterium.
