Figure 2.
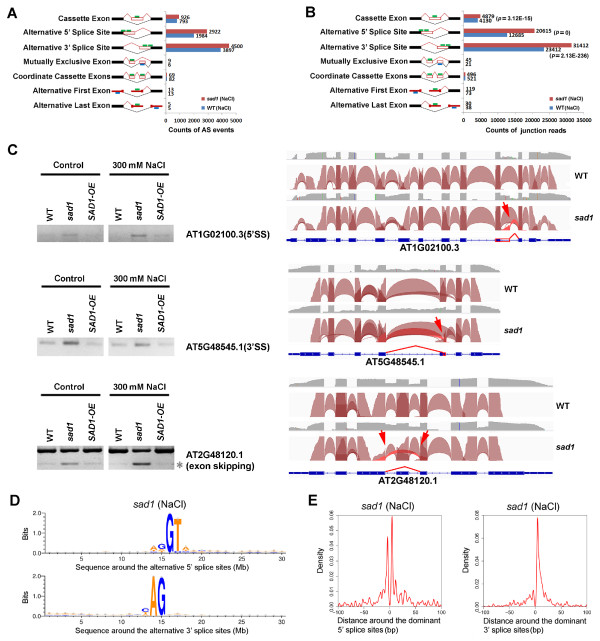
Comparison of global alternative splicing between the wild type and the sad1 mutant. (A) The counts of each type of AS events in the wild type and sad1. The green/blue bars represent forward and reverse sequencing reads. (B) The total counts of the splice junction reads from each type of AS in the wild type and sad1. The P values were calculated by Fisher’s exact test comparing the junction read counts and the uniquely mapped reads between the wild type and sad1. (C) Three representative AS events validated by RT-PCR and visualized by IGV browser. For the validation of alternative 5′SSs and 3′SSs, there was only one band that represented the alternative-splice isoform, which was obviously detected in sad1 mutants, but not in the wild type and SAD1-OE. For exon-skipping events, the grey asterisk (*) to the right side denotes the alternative splice form. For the IGV visualization, exon-intron structure of each gene was given at the bottom of each panel. The arcs generated by IGV browser indicate splice junction reads that support the junctions. The grey peaks indicate RNA-seq read-density across the gene. The upper, middle and lower panels show the indicated genes with alternative 5′SSs, alternative 3′SSs and exon-skipping, respectively. These events were marked by red arrows and highlighted by red arcs. (D) The sequences around the alternative 5′SSs and 3′SSs that were over-represented in the mutant are shown by Weblogo. (E) Distribution of activated alternative 5′SSs and 3′SSs around the dominant ones. These alternative 5′SSs and 3′SSs were enriched in the downstream or upstream 10 bp region of the dominant 5′SSs and 3′SSs (position 0 on the x- axis), respectively. AS, alternative splice; sad1, sad1 mutant; SAD1-OE, plants over-expressing wild-type SAD1 in the sad1 mutant background; WT, wild type.
