Figure 1.
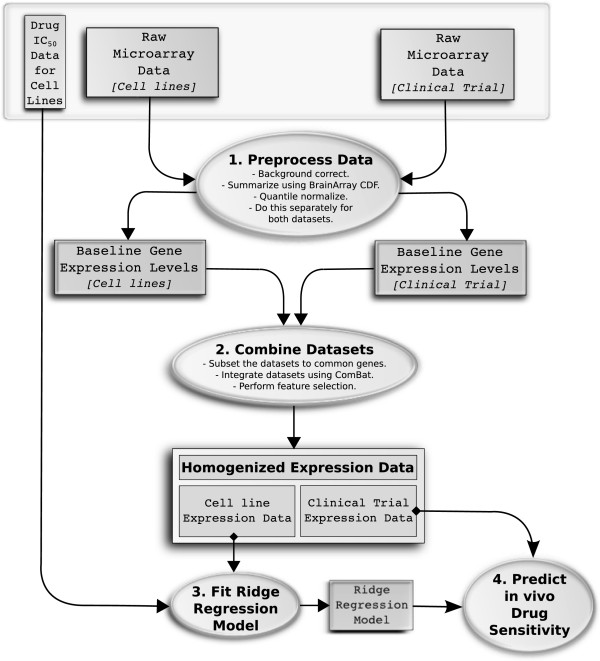
Data flow diagram showing our approach to predicting in vivo drug sensitivity. Data are represented by rectangles and processes by ovals. The input data (baseline expression and drug IC50 in cell lines and in vivo tumor gene expression) are shown in a gray box. The raw microarray data are (1) preprocessed separately using the robust multi-array average [26] method and the CDF files remapped by BrainArray [27] are summarized, (2) then combined and homogenized using ComBat. (3) A ridge regression model is fitted for baseline gene expression levels in the cell lines against the in vitro drug IC50 estimates and (4) this model is then applied to the baseline tumor expression data from the clinical trial, to yield drug sensitivity estimates. Complete details are given in Materials and methods. CDF, chip definition file.
