Figure 2.
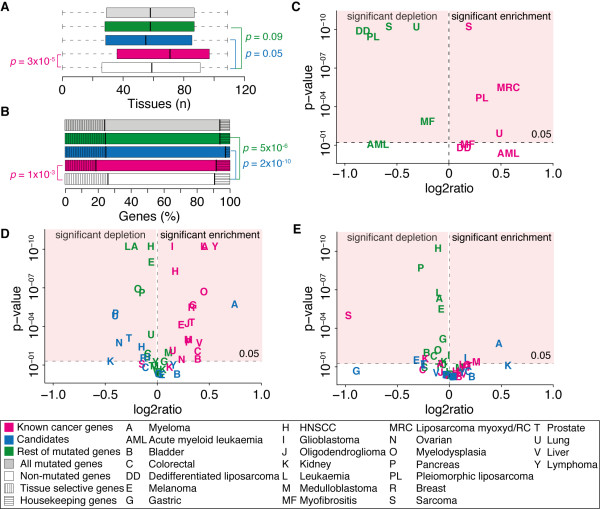
Expression of known, candidate and rest of mutated genes in cancer and normal tissues. (A) Breadth of expression of mutated genes in healthy tissues. Since the data were not normally distributed (P value 10-42, Shapiro-Wilk test, Additional file 1, Figure S1), distributions were compared using the Wilcoxon test. (B) Fraction of housekeeping and tissue-specific mutated genes. Housekeeping genes were defined as genes expressed in 107/109 tissues (98%). Tissue specific genes were defined as genes expressed in 27/109 tissues (<25%). Fisher's exact test with one degree of freedom was used to determine statistical significance. (C) Volcano plot showing the log2ratios between the fractions of expressed genes in each group of mutated genes and in non-mutated genes. For each log2ratio, the corresponding P value from the chi-squared test with one degree of freedom is also shown. None of the three studies used for this analysis [36,43,44] identified candidate cancer genes, thus only the expression of known cancer genes and other mutated genes could be checked. (D) Volcano plot showing the log2ratios between the fractions of mutated genes (known cancer genes, candidates and other mutated genes) and non-mutated genes that are expressed in the normal counterparts of the 20 tumour types. The P value from the chi-squared test, one degree of freedom for each log2ratio is also shown. For assignment of normal tissues to tumour types see Additional file 2, Table S3. (E) Volcano plot showing the log2ratios of the faction of highly expressed mutated genes compared with the rest of highly expressed human genes. Highly expressed genes were identified as those genes with expression higher than the median expression for that tissue (see Methods).
