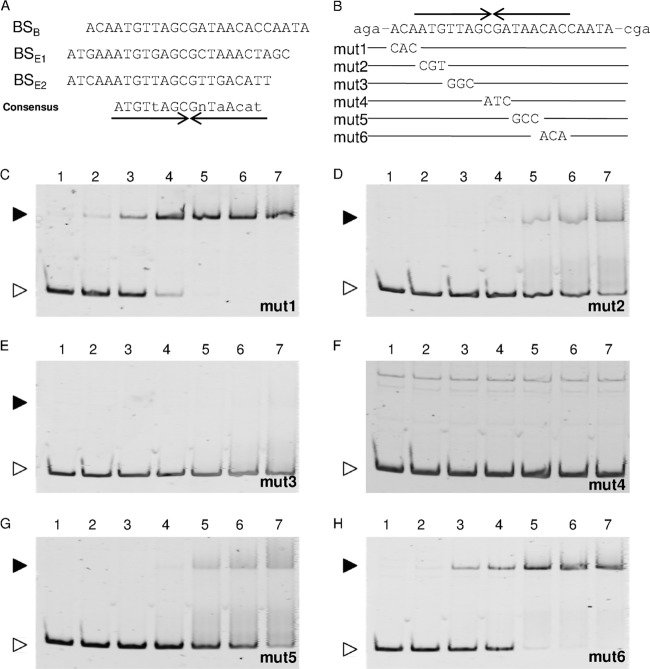
FIG 9.
The AraR-binding motif. (A) The three AraR-binding sites determined by DNase I footprinting analyses, located in the araB (BSB) and araE (BSE1 and BSE2) upstream regions, are aligned with the consensus sequence, in which the nucleotides in capital and lowercase letters are identical in all and two of the three sites, respectively, and “n” stands for any nucleotide. Various mutations (mut1 to mut6) were introduced into the AraR-binding site within the araB upstream region (B) and used for EMSAs (C to H). An inverted repeat sequence is indicated by arrows under (A) or above (B) the sequence. Each 20-μl binding reaction mixture, containing the DNA probe at 1 nM and the AraR protein at various concentrations (lanes 1, no protein; lanes 2, 10 nM; lanes 3, 25 nM; lanes 4, 50 nM; lanes 5, 75 nM; lanes 6, 100 nM; and lanes 7, 125 nM), was subjected to electrophoresis on a 6% polyacrylamide gel. The free DNA and the DNA-protein complex are indicated by white and black arrowheads, respectively.