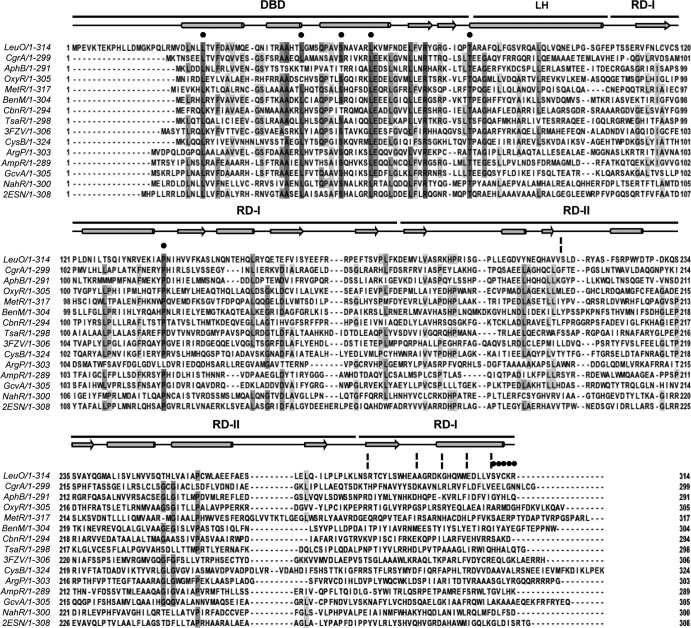
FIG 3.
Multiple-sequence alignment of LeuO and several characterized LTTRs. LeuO has a 20-residue extension at the N terminus when its sequence is aligned with the sequences of the other LTTRs. The DNA-binding domain of LeuO comprises residues 21 to 78, the linker helix comprises residues 79 to 105, and the C-terminal regulatory domain comprises residues 106 to 314. Black circles and broken vertical lines highlight the replaced residues and deletions, respectively. The alignments and prediction of the secondary structure (in which the schematic representation above the sequences shows helices as cylinders, β sheets as arrows, and turn segments as lines) of LeuO were performed with the ClustalW and JalView programs (59). Dark gray and pale gray boxes show residues with the highest and the least identity, respectively. LysR family domains DBD, LH, and the C-terminal regulatory domain with two subdomains, RD-I (central, C terminal) and RD-II, are shown at the top. The amino acid sequences used in the alignment are as follows: LeuO (S. enterica serovar Typhi), CgrA (Neisseria meningitidis, GI 7188597), AphB (V. cholerae, GI 5565924), OxyR (E. coli, GI 388479299), MetR (E. coli, GI 388479422), BenM (Acinetobacter baylyi ADP1, GI 2996626), CbnR (Ralstonia eutropha, GI 4210464), TsaR (Comamonas testosteroni T-2, GI 75499536), CysB (E. coli, GI 1787530), ArgP (Mycobacterium tuberculosis H37v, GI 15609122), AmpR (Citrobacter freundii, GI 736669), GcvA (E. coli, GI 388478824), and NahR (Pseudomonas putida, GI 37220701). The PDB accession number for P. aeruginosa Q9I6S0 is 3FZV, and that for P. aeruginosa Q9I641 is 2ESN.