Abstract
The assembly of the bacterial flagellum is exquisitely controlled. Flagellar biosynthesis is underpinned by a specialized type III secretion system that allows export of proteins from the cytoplasm to the nascent structure. Bacillus subtilis regulates flagellar assembly using both conserved and species-specific mechanisms. Here, we show that YvyG is essential for flagellar filament assembly. We define YvyG as an orthologue of the Salmonella enterica serovar Typhimurium type III secretion system chaperone, FlgN, which is required for the export of the hook-filament junction proteins, FlgK and FlgL. Deletion of flgN (yvyG) results in a nonmotile phenotype that is attributable to a decrease in hag translation and a complete lack of filament polymerization. Analyses indicate that a flgK-flgL double mutant strain phenocopies deletion of flgN and that overexpression of flgK-flgL cannot complement the motility defect of a ΔflgN strain. Furthermore, in contrast to previous work suggesting that phosphorylation of FlgN alters its subcellular localization, we show that mutation of the identified tyrosine and arginine FlgN phosphorylation sites has no effect on motility. These data emphasize that flagellar biosynthesis is differentially regulated in B. subtilis from classically studied Gram-negative flagellar systems and questions the biological relevance of some posttranslational modifications identified by global proteomic approaches.
INTRODUCTION
The bacterial flagellum is a complex molecular motor that has been shown to play roles in motility, surface adherence, biofilm structure, and signal transduction (1–4). The flagellum is organized into three main structural components: the basal body, hook, and filament (1). The basal body consists of the flagellar motor, which is required to power rotation of the flagellum, and a type III secretion (T3S) system that permits the export of proteins required for the biosynthesis of the hook and filament. The hook is a flexible joint that permits a change in the angle of rotation of the flagellum, while the filament acts as a propeller to drive movement. Biosynthesis of the flagellum is tightly regulated at the level of transcription. In the Gram-positive bacterium Bacillus subtilis, the proteins needed for the hook-basal body (HBB) are transcribed in the 31-gene fla-che operon (5, 6). The penultimate gene of this operon, sigD, encodes the sigma factor σD (7, 8) that activates transcription of the late flagellar genes: the flagellar filament gene hag; the flagellar stator genes motA and motB; the anti-sigma factor flgM; the hook-filament junction genes flgK and flgL (9); and the autolysins (10). In wild-type B. subtilis, while all cells transcribe the fla-che operon, only a subpopulation of the cells synthesize flagella (11, 12). This is due to heterogeneity in sigD transcription such that a threshold level of sigD transcription must be reached to allow sufficient σD protein to accumulate and activate σD-regulated promoters (11). The net result is that transcription is temporally ordered such that the HBB genes are expressed before the filament genes (1).
As well as being controlled at the level of transcription, flagellar biosynthesis is regulated posttranscriptionally by flagellar type III secretion system (T3S) chaperones. Regulation at this level has been described most extensively in Salmonella enterica serovar Typhimurium, with little known about the function of chaperones in Gram-positive bacterial species (13, 14). T3S chaperones are small proteins that bind their cognate substrate(s) in the cytoplasm, protecting the substrate from degradation and/or preventing aggregation (15, 16). Chaperones therefore allow the efficient transport of the substrate to the export machinery. Chaperone-substrate complexes reach the secretion apparatus with the aid of the soluble export apparatus proteins FliI and FliH (17), while empty chaperones are recycled with the aid of FliJ (18). Following interaction of the chaperone-substrate complex with the C-terminal cytoplasmic domain of the integral membrane protein FlhA, a series of protein-protein interactions facilitates the entry of the substrate protein to the export gate (19–21), and its subsequent secretion is driven by proton motive force (22, 23). In S. Typhimurium, FliS is a specific chaperone for flagellin (24), FliT is specific for the FliD filament cap protein, and FlgN (ST-FlgN) is specific for the hook-filament junction proteins, FlgK and FlgL (25, 26). Recently, it has been shown that FliS is required for Hag (flagellin) secretion in B. subtilis (13, 14). In addition, in silico analysis has suggested that YvyG of B. subtilis is an orthologue of the S. Typhimurium protein FlgN (27). However, a defined function for YvyG has not yet been determined experimentally.
Several global proteomic screens have been conducted using B. subtilis with the goal of examining the extent and diversity of posttranslational modification (28–30). Intriguingly, these experiments identified YvyG as being phosphorylated on tyrosine 49 (29) and arginine 60 (30). Posttranslational modification of proteins can control cell fate in several ways: (i) by altering protein localization and half-life (31), (ii) by controlling protein activity and affinity to ligands (32), and (iii) by the disruption or promotion of protein-protein interactions (33). The B. subtilis flagellum has recently been shown to be regulated by mechanisms not identified in other bacterial species (34, 35). Therefore, given the potential for YvyG to play a crucial role in the tightly controlled process of flagellar biosynthesis (31, 36), we hypothesized that protein phosphorylation might present an additional route for B. subtilis to regulate flagellar assembly and, therefore, motility. Thus, we aimed to define the function of YvyG during motility by B. subtilis and to ascertain the in vivo role of YvyG tyrosine and arginine phosphorylation.
Work presented here identifies B. subtilis YvyG (here referred to as B. subtilis FlgN [Bs-FlgN]) as an orthologue of the S. Typhimurium T3S chaperone FlgN, as previously suggested by in silico analysis (27). Consistent with this, we prove for the first time that FlgN is required for both swimming and swarming motility in B. subtilis. The lack of motility in the B. subtilis flgN deletion strain is linked to a block in flagellar biosynthesis. This is a consequence of a complete lack of filament assembly. Analysis of a ΔflgK-flgL double mutant strain demonstrates that the strain phenocopies the flgN mutation. Collectively, these data prove that in B. subtilis FlgN is required for flagellar assembly, perhaps by acting as a chaperone for FlgK and FlgL. In S. Typhimurium, overexpression of flgK can compensate for the motility defect of a ΔflgN strain (16). However, data presented here demonstrates that in B. subtilis deletion of flgN cannot be compensated for by the overexpression of flgK and flgL. This leads to the conclusion that there is a stricter dependence on the presence of FlgN in B. subtilis for motility than there is for FlgN in S. Typhimurium (16, 36). Finally, through the use of site-directed mutagenesis, we demonstrate that mutation of the tyrosine and arginine phosphorylation sites of FlgN has no effect on the ability of B. subtilis to become motile. In summary, these data emphasize that flagellar biosynthesis is differentially regulated in B. subtilis in comparison to the classically studied Gram-negative bacteria and additionally raises questions regarding the biological relevance of some posttranslational modifications identified by global proteomic approaches.
MATERIALS AND METHODS
Growth conditions and strain construction.
Escherichia coli and Bacillus subtilis strains were routinely grown in Luria-Bertani (LB) medium (10 g liter−1 NaCl, 5 g liter−1 yeast extract, 10 g liter−1 tryptone) or on LB plates supplemented with 1.5% agar at 37°C unless otherwise stated. E. coli strain MC1061 [F′ lacIq lacZΔM15 Tn10(tet)] was used for the routine construction and maintenance of plasmids. When required, antibiotics were used at the following concentrations: 100 μg ml−1 ampicillin, 100 μg ml−1 spectinomycin, 25 μg ml−1 chloramphenicol, 10 μg ml−1 kanamycin (B. subtilis), 50 μg ml−1 kanamycin (E. coli), 1 μg ml−1 erythromycin, and 25 μg ml−1 lincomycin. Strains were constructed using standard protocols. Phage transductions were carried out as previously described (37). When appropriate, isopropyl β-d-1-thiogalactopyranoside (IPTG) was added at the concentrations indicated in the figures or figure legends. A full list of strains used in this study is provided in Table S1 in the supplemental material.
Construction of in-frame deletion strains.
To construct the in-frame deletion of flgN, an approach similar to that previously described was used (38). The upstream region of flgN was amplified from genomic DNA using primers NSW938 and NSW939, purified, and digested with XbaI and SalI using the restriction sites engineered into the primers. The downstream region of flgN was amplified using primers NSW936 and NSW937, purified, and digested with BamHI and XbaI using the restriction sites engineered into the primers. The fragments were simultaneously ligated into pUC19 and sequenced, prior to introduction into pMAD (39), to produce plasmid pNW399. Strain NRS3570 (NCIB3610 ΔflgN) was generated by integration and curing of the region contained in pNW399 in strain NCIB3610. Strains NRS4041 (NCIB3610 ΔfliD), NRS4042 (NCIB3610 ΔflgE), and NRS4060 (NCIB3610 ΔflgK-flgL) were constructed in a similar manner using the primers and plasmids detailed in Tables S2 and S3 in the supplemental material, respectively, and in the supplemental methods.
Introduction of site-specific mutations to the chromosome.
To introduce site-specific mutations to the chromosome of NCIB3610, a similar approach to that previously described was used (39). Plasmids pNW801 and pNW1012 were used to introduce site-specific mutations in codon 49 of flgN. Primers NSW936 and NSW939 were used to amplify a 1,725-bp region of DNA containing the complete flgN coding region. The PCR fragment was cloned into pUC19 using SalI and SphI restriction sites engineered into the primers, resulting in plasmid pNW398. Primer sets NSW942 and NSW943 (Y49A) and NSW1436 and NSW1437 (Y49E) were used to introduce point mutations to pNW398 using site-directed mutagenesis by the QuikChange method, according to the manufacturer's instructions (Stratagene). The resulting plasmids were sequenced to ensure that the correct mutations were introduced. The mutated flgN coding region was excised and cloned into pMAD for integration into NCIB3610, as described for the construction of in-frame deletion strains. Further plasmids were constructed in an identical manner for hag (Hag T209C) and flgN (with the R-to-A or R-to-E mutation at position 60 encoded by flgN [flgN-R60A or flgN-R60E, respectively]). Full details of the plasmids and primers used are provided in Tables S2 and S3, respectively, in the supplemental material.
Sigma A antibodies.
To overexpress and purify the σA protein from B. subtilis for antibody preparation, the sigA gene was amplified from the chromosome of the strain NCIB3610 using primers NSW860 and NSW861. The NcoI and XhoI sites engineered into the primer sequence are underlined in Table S3 in the supplemental material. The resulting PCR product was digested with NcoI and XhoI and cloned into the expression vector pEHISGFPTEV (40) to yield a His6-green fluorescent protein (GFP)-σA fusion construct named pNW642. A tobacco etch virus (TEV) protease recognition site was placed between the gfp and sigA coding regions.
E. coli BL21(DE3) cells carrying the pNW642 vector were grown in LB broth containing ampicillin to an optical density at 600 nm (OD600) of 0.3 at 37°C. The cells were chilled to 20°C, and protein expression was induced with 50 μM IPTG overnight. Cells were collected by centrifugation and resuspended in lysis buffer (25 mM Tris [pH 7.5], 250 mM NaCl, 5 mM dithiothreitol [DTT], 30 mM imidazole, lysozyme, DNase I, and complete EDTA-free protease inhibitor cocktail [Roche]). Cells were lysed on a French press using pressure at 15,000 lb/in2, and the cellular debris was removed by centrifugation. The supernatant was filtered through a 0.45-μm-pore-size syringe filter before being loaded onto a 1-ml HiTrap HF immobilized metal affinity chromatography (IMAC) column (GE Healthcare) using loading buffer, and the column was then washed with 10 ml of loading buffer (25 mM Tris [pH 7.5], 250 mM NaCl, 5 mM DTT, 30 mM imidazole). The recombinant His6-GFP-σA fusion protein was eluted from the column using a gradient of elution buffer (25 mM Tris [pH 7.5], 250 mM NaCl, 5 mM DTT, 500 mM imidazole). The fractions containing the fusion protein were pooled and dialyzed into TEV buffer (50 mM Tris [pH 7.5], 20 mM NaCl, 0.5 mM EDTA, 10% glycerol) using spin concentrators. The dialyzed protein was diluted to 1 mg ml−1 in the TEV buffer, and 1.5 mg of TEV protease was added. The reaction solution was incubated overnight at 4°C with agitation. The resulting σA and GFP proteins were separated using negative IMAC with the loading and elution buffers described above. The unbound fraction, containing σA, was additionally purified using size exclusion chromatography with a Superdex75 resin (GE Healthcare) and buffer containing 25 mM Tris (pH 7.5) and 250 mM NaCl. The purified protein was concentrated to 1 mg ml−1 and sent for rabbit immunization to Dundee Cell Products (Dundee, United Kingdom). The obtained σA antiserum was affinity purified against purified recombinant σA according to a previously described protocol (41).
Secondary-structure prediction.
Primary protein sequences of S. Typhimurium FlgN and B. subtilis FlgN were aligned using Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) (42). The secondary structures were predicted using PsiPred (http://bioinf.cs.ucl.ac.uk/psipred/) (43, 44) and aligned against the primary sequence of the proteins.
RNA extraction and RT-PCR.
RNA isolation was carried out as described previously (38) using a RiboPure Bacteria RNA Isolation Kit (Ambion), according to the manufacturer's instructions, and treated with DNase I. To confirm cotranscription of flgN with flgK and flgL, cDNA was synthesized using the yviE gene-specific primer NSW1459 in a reaction with SuperScript III (Life Technologies) and subsequently treated with RNase H (NEB) for 20 min at 37°C. To establish if deletion of flgN perturbed transcription of flgK or flgL (and vice versa), cDNA was synthesized using random hexamers in a reaction with SuperScript III (Life Technologies). To amplify internal gene products, the following primer pairs were used: DEN5 and DEN7 (rRNA), NSW1446 and NSW1447 (flgL), NSW1444 and NSW1445 (flgK), NSW1442 and NSW1443 (flgN), and NSW1440 and NSW1441 (flgM).
Motility assays.
Swimming and swarming analyses were performed as described previously (37) using low-salt LB medium (5 g liter−1 NaCl, 5 g liter−1 yeast extract, 10 g liter−1 tryptone) supplemented with 0.4% and 0.7% Bacto agar, respectively. Plates were incubated at 37°C, and the extent of swimming or swarming was noted at defined time intervals.
Staining of flagella and fluorescence microscopy.
Cells carrying the Hag T209C point mutation (45) were grown to mid-exponential phase, and 0.5 ml of cells was harvested by centrifugation at 4,000 × g. Cells were washed once with 1× T-Base [1 mM EDTA, 15 mM (NH4)2SO4, 80 mM K2HPO4, 44 mM KH2PO4, 3.4 mM sodium citrate], pelleted, resuspended in 50 μl of T-Base containing 5 μg/ml Alexa Fluor 488 C5 maleimide dye (Molecular Probes), and incubated for 5 min at room temperature. Cells were washed three times with 500 μl of 1× T-Base and suspended in 50 μl of 1× phosphate-buffered saline (PBS). Two microliters of the cell suspension was spotted onto a thin matrix of 1.5% agarose in water (Ultrapure Agarose; Invitrogen) contained in a 1.7- by 2.8-cm Gene Frame (AB-0578; ABgene House, Epsom, Surrey, United Kingdom) mounted on a standard microscope slide (Super Premium slides; VWR). Each slide was prepared as follows: the gene frame was filled with molten 1.5% agarose and covered firmly with a standard microscope slide to flatten the agarose surface. Following solidification of the agarose, the slide was carefully removed, and the cell suspension was added. Once the cell suspension was dry, the gene frame was sealed with a coverslip (thickness number 1.5; VWR), and images were immediately acquired. Imaging was performed using a DeltaVision Core wide-field microscope (Applied Precision) mounted on an Olympus lX71 inverted stand with an Olympus 100× (1.4 numerical aperture [NA]) lens and a CoolSNAPHQ camera (Photometrics) with differential interference contrast (DIC) and fluorescence optics. GFP was imaged using a 100 W Mercury lamp and a fluorescein isothiocyanate (FITC) filter set (excitation, 490/20 nm; emission, 528/38 nm) with an exposure time of 200 ms. DIC images were illuminated with an LED-transmitted light source.
To monitor Phag-yfp expression, cells were grown at 37°C in LB medium to an OD600 of 1.0, 0.5 ml of the culture was harvested, and cells were washed and resuspended in 1× PBS. The cell suspension was prepared for microscopy and imaged as described above. Yellow fluorescent protein (YFP) fluorescence was imaged using a 100 W Mercury lamp and an FITC filter set (excitation, 490/20 nm; emission, 528/38 nm) with an exposure time of 50 ms. The threshold used to define activation of the transcriptional reporter Phag was set as a YFP fluorescence intensity value greater than 2 standard deviations above the mean background fluorescence. All images were rendered and analyzed postacquisition using OMERO software (www.openmicroscopy.org) (46).
Flow cytometry analysis.
The fluorescence of strains harboring yfp or gfp transcriptional promoter fusions was measured in single cells extracted from planktonic cultures grown to mid-exponential phase and analyzed as described previously (47).
Whole-cell analysis of Hag.
Proteins were extracted from planktonic cultures grown to mid-exponential phase. Briefly, cells were harvested by centrifugation at 4,700 × g. Cells were suspended in 1× Bugbuster (Novagen) and lysed according to the manufacturer's instructions. Seven micrograms of protein was resolved by SDS-PAGE and stained with Coomassie brilliant blue. Hag was identified by comparison with the Δhag strain (DS1677) and confirmed by mass spectrometry (FingerPrints Proteomics and Mass Spectrometry Facility, University of Dundee).
Western blot analysis.
Cellular proteins were extracted as for whole-cell analysis of Hag. Extracellular proteins were extracted from the culture supernatant and processed as detailed previously (48) and suspended in 50 μl of 4× SDS loading dye. Seven micrograms of cellular proteins or 7 μl of extracellular proteins was separated by SDS-PAGE prior to transfer onto polyvinylidene difluoride (PVDF) membrane (Millipore) by electroblotting. Antibodies raised against Hag (a kind gift from Kürsad Turgay) were used at 1:40,000, anti-FlgE (a kind gift from Daniel Kearns) was used at 1:20,000, anti-σA was used at 1:500, and goat anti-rabbit or goat anti-mouse horseradish peroxidase (HRP)-conjugated secondary antibodies (both from Pierce) were used at 1:5,000.
β-Galactosidase assays.
The β-galactosidase activity of strains harboring lacZ promoter reporter fusions was measured as previously described (37, 49). The values presented are the average β-galactosidase activities in Miller units (50) determined from at least three independent samples. Error bars represent the standard errors of the means.
Enrichment of flagellar hook-basal bodies.
The flagellar HBB fraction of NCIB3610 was enriched for as described previously (51, 52). Briefly, 1 liter of cells was grown to early exponential phase and harvested by centrifugation at 6,000 × g for 45 min at 4°C. The cell pellet was resuspended in 100 ml of sucrose solution (0.5 M sucrose, 0.15 M Tris) with a protease inhibitor tablet (Roche), and cells were homogenized with a loose pestle on ice. To lyse cells and allow spheroplast formation, lysozyme was added to the cell suspension at a final concentration of 0.1 mg ml−1, and samples were incubated at 4°C with stirring for 40 min. Spheroplasts were lysed by addition of Triton X-100 to a final concentration of 1%, and viscosity was decreased by stirring at room temperature for 30 min, allowing endogenous DNases to degrade cellular DNA. Unlysed cells were removed by centrifugation at 4,000 × g for 10 min, and EDTA was added to the suspension at a final concentration of 10 mM. To aid removal of contaminating membrane proteins, the pH of the lysate was raised to 10 by addition of NaOH. The lysate then underwent high-speed centrifugation (60,000 × g for 60 min), and the pellets were resuspended in alkaline solution (0.1 M KCl-KOH, 0.5 M sucrose, 0.1% Triton X-100, pH 11.0) and centrifuged again. The pellet was then resuspended in 90 ml of TET buffer (10 mM Tris-HCl, 5 mM EDTA, 0.1% Triton X-100, pH 8.0), and a 36% CsCl gradient was established. The solution was centrifuged using a Beckman SW41Ti swinging-bucket rotor at 55,000 × g for 16 h at 15°C. The flagellar fraction was visible as a band approximately 2 cm from the bottom of the tube and was collected with a Pasteur pipette and dialyzed against TET buffer. In an attempt to dissociate the flagellar filaments, the flagellar fraction was suspended in acidic solution (glycine-HCl, 0.1% Triton X-100, pH 3.0) for 1 h, and HBB complexes were collected by centrifugation at 100,000 × g for 1 h. HBBs were resuspended in TET buffer and analyzed by mass spectrometry. Results were searched against the Bacillus subtilis Mascot database (FingerPrints Proteomics and Mass Spectrometry Facility, University of Dundee). The top 12 proteins as identified by their Mascot scores are listed in Table S4 in the supplemental material.
RESULTS
YvyG of B. subtilis shares secondary-structure homology with the Salmonella protein FlgN.
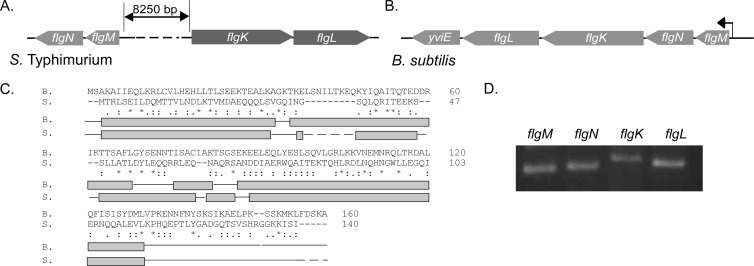
Previous bioinformatic analysis postulated that YvyG of B. subtilis was an orthologue of FlgN (27). FlgN is an essential component of the flagellar type III secretion machinery (26, 36, 53). FlgN has been most extensively studied in S. Typhimurium (26, 36), but homologues have been recognized in a number of different bacterial species (27, 53). Three criteria were employed by Pallen et al. (27) to identify FlgN homologues in a broad range of bacterial species: (i) the protein must be encoded by a gene adjacent to an flgM homologue (Fig. 1A and B), (ii) the gene should be of a similar length to flgN of Salmonella (Fig. 1C), and (iii) the protein must be recognizable (however distantly) by PSI-BLAST analysis as “FlgN-like.” These criteria were set as the primary amino acid sequence of FlgN is highly variable (27). To confirm that YvyG is likely to be an orthologue of FlgN, the amino acid sequence of YvyG was compared with that of S. Typhimurium FlgN. As expected, there was little primary-sequence homology (Fig. 1C). However, upon comparison of the predicted secondary structures of YvyG and FlgN, a high degree of similarity was clearly apparent (Fig. 1C). Thus, YvyG was renamed FlgN, and its function in motility in B. subtilis was further investigated.
FIG 1.
Primary-sequence and secondary-structure comparison of B. subtilis FlgN with S. Typhimurium FlgN. Schematic representations of the chromosomal regions surrounding S. Typhimurium flgN (A) and B. subtilis flgN (B) are shown. Arrows represent open reading frames, with the direction of the arrow indicating the direction of the open reading frame. The bent arrow represents the promoter located before the flgM coding region. (C) Primary-sequence alignment and secondary-structure comparison of Bs-flgN and ST-flgN. The primary amino acid sequence of FlgN from B. subtilis 168 (B) was aligned with that of flgN from S. Typhimurium (S). For primary-sequence alignments, an asterisk indicates a fully conserved amino acid, a colon indicates a highly conserved amino acid, and a period indicates a weakly conserved amino acid. Gaps indicate no homology. Dashed lines indicate a break in the sequence. For secondary-structure alignments light gray boxes indicate α-helices, solid lines indicate coiled coils, and dashed lines indicate a break in the sequence alignment. (D) RT-PCR analysis of cotranscription of flgM, flgN, flgL, and flgK. Regions of DNA internal to flgM, flgN, flgK, and flgL were amplified from cDNA generated using a primer specific to the gene 3′ proximal to flgL, using RNA from the wild-type strain (NCIB3610).
On the B. subtilis chromosome, the flgM, flgN, flgK, and flgL coding regions are adjacent. To ascertain whether all four genes were part of the same operon in the NCIB3610 strain, reverse transcription-PCR analysis was used. RNA was extracted from wild-type B. subtilis, and cDNA was synthesized using a primer specific to yviE, the gene proximal to flgL at the 3′ end (Fig. 1B). The cDNA generated was used as a template for PCR with primer pairs specific to the internal coding regions of flgM, flgN, flgK, and flgL. Regions of DNA internal to the coding regions of flgM, flgN, flgK, and flgL could each be amplified from cDNA generated using a primer at the 3′ end of flgL (just beyond the termination codon) (Fig. 1D), demonstrating that all four genes are cotranscribed.
FlgN is essential for swarming and swimming motility of B. subtilis.
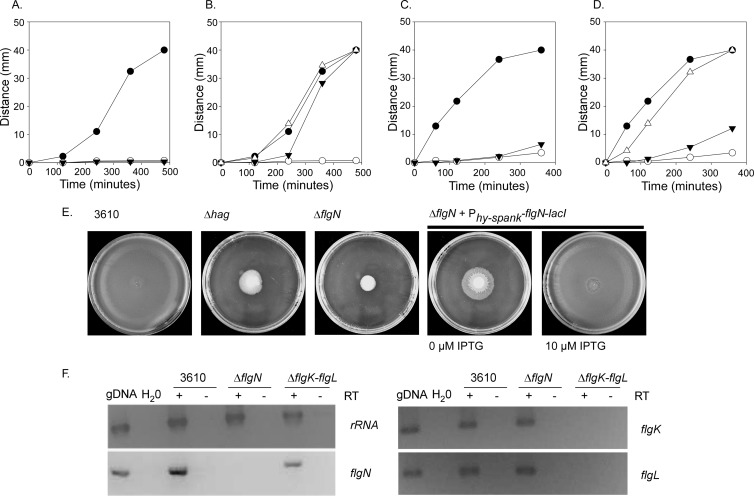
The flgN gene is located in a region of the chromosome known to be required for motility (9). To test if flgN is required for motility, a B. subtilis strain carrying an in-frame deletion of flgN was constructed, and its phenotype was assessed in swimming and swarming assays. A strain containing a deletion in the hag gene (DS1677) was used as a nonmotile control. While the wild-type strain was able both to swim and swarm efficiently (Fig. 2A, C, and E), this behavior was lost in the ΔflgN strain (NRS3570) (Fig. 2A, C, and E). Using reverse transcription-PCR analysis, we confirmed that transcription of the other genes in the operon was not impacted by the flgN deletion (Fig. 2F). To further ensure that this loss of motility was specific to the deletion of flgN, the motility of a strain where the coding region of flgN was reintroduced at the amyE locus under the control of an IPTG-inducible promoter (Phy-spank) was tested. Both the swimming (Fig. 2B) and swarming (Fig. 2D and E) phenotypes of the ΔflgN strain could be restored by the reintroduction of flgN on the chromosome upon induction with IPTG. These data demonstrate for the first time in vivo that the protein product of the flgN gene is required for both swimming and swarming motility in B. subtilis. It is notable that higher levels of flgN transcription are required to complement the swarming defect presented by the ΔflgN strain than are needed for the swimming defect (compare Fig. 2B and D). This is most likely attributable to a higher demand for flagellum biosynthesis in swarming than swimming motility (54).
FIG 2.
ΔflgN strains are nonmotile. Swim expansion assays (A and B) and swarm expansion assays (C and D) were performed. Wild-type (3610; filled circle), Δhag (DS1677; filled triangle), and ΔflgN (NRS3570; open circle) strains were used for the experiments shown in panels A and C. Wild-type (3610; filled circle) and ΔflgN (NRS3570; open circle) strains along with the ΔflgN amyE::Phy-spank-flgN-lacI (NRS3578) strain without (filled triangle) and with (open triangle) 10 μM IPTG induction were used for the experiments shown in panels B and D. Each graph is representative of three independent biological replicates. (E) Photographs of swarm expansion plates taken at the end of the assay, after 6 h of incubation at 37°C. (F) RT-PCR analysis of transcription of rRNA, flgN, flgK, and flgL in wild-type (NCIB3610), ΔflgN (NRS3570), and ΔflgK-flgL (NRS4060) strains. Genomic DNA (gDNA) and H2O are shown as positive and negative controls for amplification, respectively. Reaction mixtures were incubated with (+) or without (−) reverse transcriptase (RT).
Deletion of flgN results in a loss of bimodal transcription of hag.
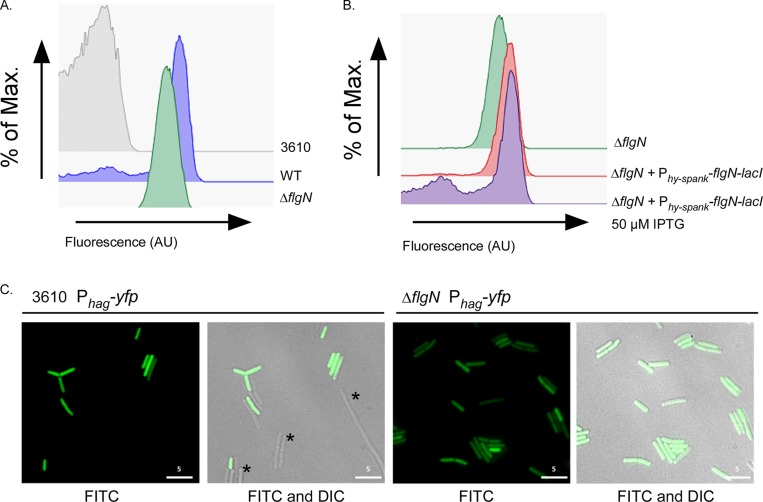
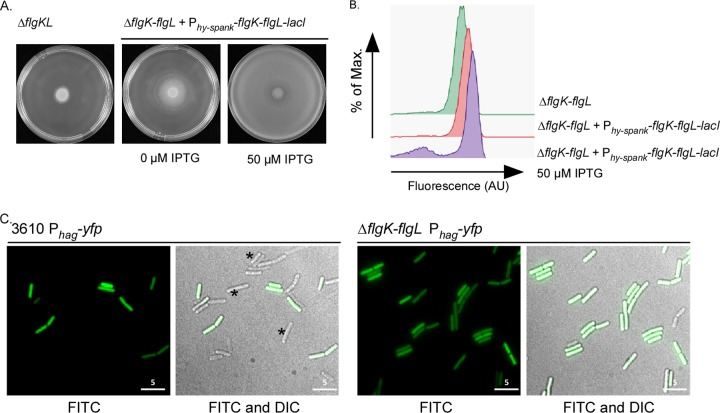
To dissect the role of flgN in motility, the effect of deletion of flgN on the transcription of the flagellar filament gene hag was tested. Due to the heterogeneity in sigD transcription, transcription of hag is bimodal, and thus single-cell techniques are ideally suited for analysis (11, 12). To this end, a Phag-yfp transcriptional reporter was integrated at a heterologous location on the chromosome. Flow cytometry and single-cell microscopy were used to assess the transcription profile using the fluorescence generated by YFP as a reporter. In the wild-type strain, the bimodality of hag transcription in the cell population is clearly evident (Fig. 3A and C). Strikingly, upon deletion of flgN bimodality is lost as hag transcription was observed for all cells within the population, albeit at a slightly lower level than in the wild type (Fig. 3A and C). The alteration in the hag transcription profile can be complemented upon reintroduction of flgN under the control of an IPTG-inducible promoter at the heterologous amyE locus, confirming the requirement of flgN for bimodal hag transcription (Fig. 3B). However, as the hag gene is still transcribed in the absence of flgN, these findings indicate that lack of motility in the ΔflgN strain is not due to a lack of hag transcription.
FIG 3.
Deletion of flgN results in a loss of bimodal hag transcription. (A and B) Flow cytometry analysis of hag transcription in strains carrying the Phag-yfp transcriptional reporter fusion. Strain 3610 was used as a nonfluorescent control. Shown are the wild-type (WT; NRS3076), ΔflgN (NRS3570), and ΔflgN amyE::Phy-spank-flgN-lacI (NRS3713) strains without and with induction with 50 μM IPTG. (C) Fluorescence microscopy analysis of the wild-type (NRS3076) and ΔflgN (NRS3708) strains carrying the Phag-yfp transcriptional reporter (false-colored green). Scale bar, 5 μm. Asterisks indicate examples of cells that do not transcribe hag. Max, maximum; AU, arbitrary units.
Loss of flgN is associated with a defect in flagellar biosynthesis.
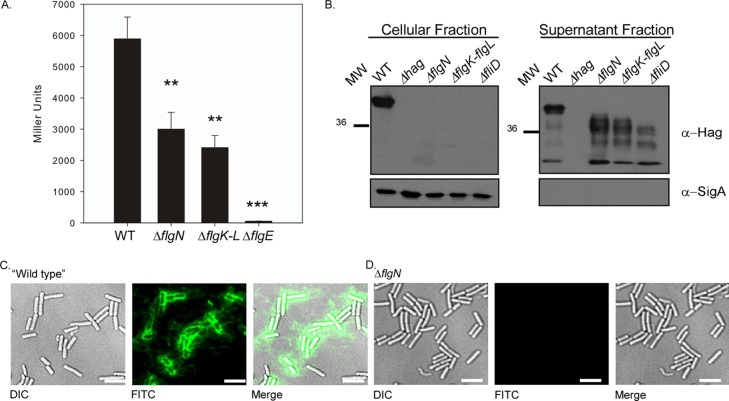
Given that the hag coding region is transcribed (Fig. 3), a lack of motility in the absence of flgN could arise from lack of translation of the hag transcript. To assess hag translation, a Phag′-′lacZ translational reporter fusion (under the control of the hag promoter, the hag leader region, Shine-Dalgarno sequence, and start codon) was constructed and introduced into B. subtilis at the amyE locus. When the β-galactosidase activity levels in the flgN mutant were compared with the wild-type levels, Hag translation was found to be 2-fold lower (Fig. 4A). However, a strain carrying an in-frame deletion of flgE, which encodes the main protein component of the flagellar hook, showed a 100-fold decrease in Hag translation (Fig. 4A) (55). It is likely that the observed decrease in the translation of hag in the ΔflgN mutant strain is a result of hag being transcribed at a slightly lower level in all cells than in the wild-type strain (Fig. 3). Alternatively, it is technically possible that translation itself may be regulated, as occurs in the absence of flgE (see Discussion) (55). While there is a statistically significant (P = 0.01) decrease in Hag translation in the absence of flgN, it did not appear to be sufficient to account for the complete lack of motility demonstrated in Fig. 2 (i.e., the severity of the motility defect does not match the small decrease in translation of Hag). A lack of motility in the presence of hag transcription and translation could be due to a lack of Hag polymerization. To test if Hag was secreted but not assembled into a flagellar filament in the ΔflgN strain, proteins were extracted from the cellular and supernatant fractions of cells grown to mid-exponential phase. The presence or absence of Hag was detected by Western blotting, with the cytoplasmic sigma factor, σA, used as a loading and fractionation control. For the wild-type strain Hag is detected in the cellular fraction (which includes assembled flagella) and in the supernatant fraction (including unassembled and sheared flagella) (Fig. 4B). However, for the ΔflgN strain Hag is present only in the supernatant fraction at a lower molecular weight (Fig. 4B). This is likely to be unpolymerized Hag or the products of proteolytic degradation resulting from the action of the extracellular proteases (13). As a positive control for Hag secretion, a strain carrying an in-frame deletion of fliD, which encodes the filament cap protein, was also assessed. As shown previously (13), this strain phenocopies the Hag secretion profile seen in the ΔflgN strain, thus supporting our conclusions.
FIG 4.
Deletion of flgN is associated with a decrease in hag translation and a block in filament assembly. (A) β-Galactosidase assays of strains carrying the Phag′-′lacZ translational reporter fusion. Shown are the wild-type (WT; NRS4795), ΔflgN (NRS4796), ΔflgK-flgL (NRS4799), and ΔflgE (NRS4798) strains. Data are plotted as the averages of at least three independent replicates. Error bars represent standard errors of the means. Asterisks denote significance as calculated by a Student t test: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (B) Western blot analysis of cellular (including assembled flagella) and supernatant (including sheared and unassembled flagella) fractions of the wild-type (3610), Δhag (DS1677), ΔflgN (NRS3570), ΔflgK-flgL (NRS4060), and ΔfliD (NRS4041) strains, separately probed with anti-Hag and anti-σA primary antibodies. MW, molecular weight in thousands; α, anti. (C and D) Fluorescence micrographs of strains carrying the Hag T209C point mutation labeled with Alexa Fluor 488 C5 maleimide (false-colored green). Shown are the wild-type (NRS3719) (C) and ΔflgN (NRS3718) (D) strains. Scale bar, 5 μm.
Loss of filament polymerization in the ΔflgN strain was confirmed by single-cell fluorescence microscopy of strains where the codon for threonine at position 209 of the hag gene was mutated to cysteine to enable labeling with an Alexa Fluor 488 C5 maleimide dye (45). In the wild-type background (NRS3719) the flagellar filaments are clearly visible (Fig. 4C). However, upon deletion of flgN, no signal was detected (Fig. 4D). Collectively, these data show that the ΔflgN strain is nonmotile due to both a small decrease in translation of Hag and a block in filament assembly, which results in the accumulation of unpolymerized Hag in the extracellular milieu. Together, these data prove that the lack of motility exhibited by the flgN mutant is due to a lack of flagellar filament assembly.
Deletion of the hook junction genes generates a strain which phenocopies the flgN mutant.
A lack of filament polymerization in the absence of flgN is consistent with the hypothesis that Bs-FlgN is an orthologue of ST-FlgN; i.e., if FlgK and FlgL are not properly localized to the hook-filament junction, the flagellar filament cannot be assembled (26, 53). We proposed that if Bs-FlgN were indeed an orthologue of ST-FlgN, then a B. subtilis strain lacking flgK and flgL might phenocopy the ΔflgN strain. A strain carrying an in-frame deletion of flgK-flgL was constructed and found to be unable to swarm (Fig. 5A). In addition, flow cytometry and microscopy analyses revealed that, like deletion of flgN, deletion of flgK-flgL resulted in a loss of bimodality with respect to transcription of the σD-regulated gene hag (Fig. 5B and C) and a 2-fold decrease in Hag translation compared with the wild-type strain (Fig. 4A). Furthermore, upon deletion of flgK-flgL, Hag could not be detected in the cellular fraction but was instead found in the extracellular milieu (Fig. 4B). Using reverse transcription-PCR analysis, we confirmed that transcription of the other genes in the operon was not impacted by the flgK-flgL deletion (Fig. 2F). All of the phenotypes were proven to be specific to deletion of the flgK-flgL coding region as the mutant strain could be complemented by replacement of the flgK-flgL coding region at a heterologous location on the chromosome under the control of the Phy-spank promoter (Fig. 5A and B). In conclusion, a double deletion of flgK and flgL generates a strain that phenocopies the flgN mutant, as demonstrated by physiological, biochemical, and single-cell analyses. This is consistent with Bs-FlgN functioning as an orthologue of ST-FlgN.
FIG 5.
The ΔflgN strain phenocopies the ΔflgK-flgL strain. (A) Photographs of swarm expansion plates taken after 6 h of incubation at 37°C. Shown are the wild-type (3610), ΔflgK-flgL (NRS4060), and ΔflgK-flgL amyE::Phy-spank-flgK-flgL-lacI (NRS 4064) strains without and with induction with 50 μM IPTG. (B) Flow cytometry analysis of hag transcription in strains carrying the Phag-yfp transcriptional reporter fusion. Shown are the ΔflgK-flgL (NRS4071) and ΔflgK-flgL amyE::Phy-spank-flgK-flgL-lacI (NRS4078) strains without and with induction with 50 μM IPTG. (C) Fluorescence microscopy analysis of the wild-type (NRS3076) and ΔflgK-flgL (NRS4071) strains carrying the Phag-yfp transcriptional reporter (false-colored green). Scale bar, 5 μm. Asterisks indicate examples of cells that do not transcribe hag.
Overexpression of flgK-flgL cannot compensate for the absence of flgN.
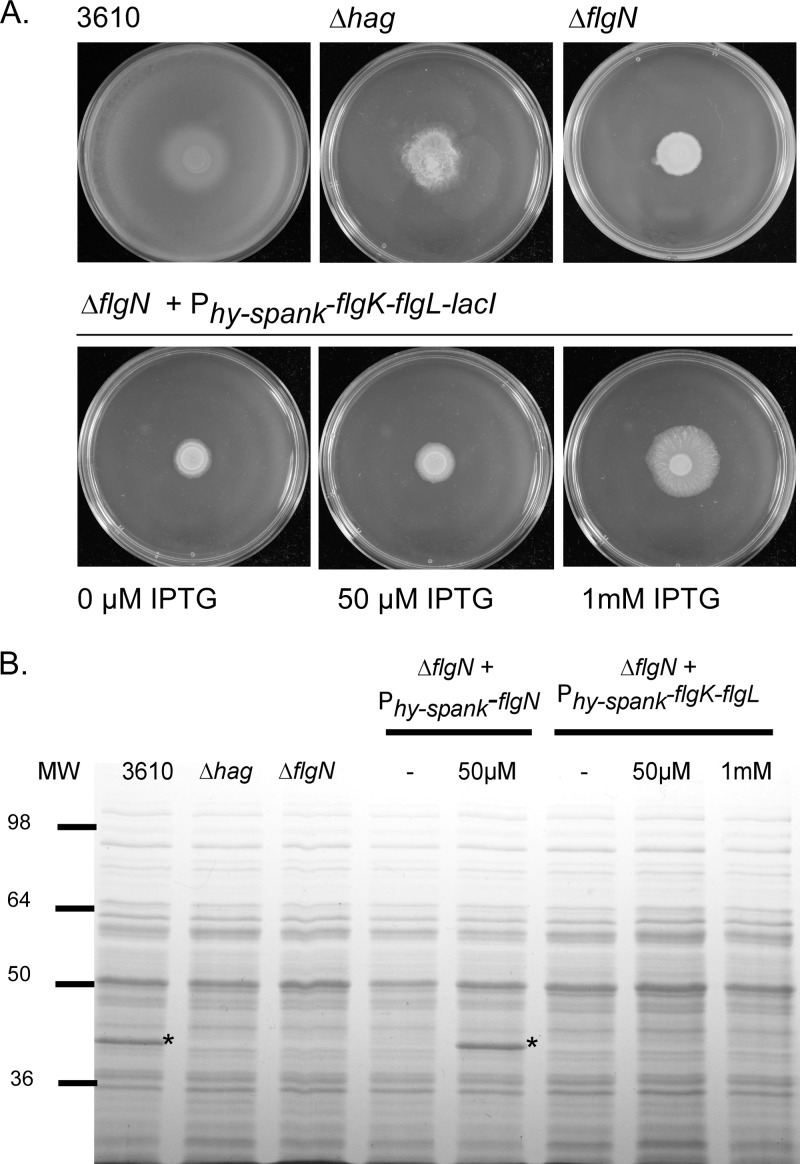
In Salmonella deletion of flgN can be compensated for by overexpression of flgK and flgL (16). This is because FlgN is not exclusively required for the secretion of its substrates but rather protects FlgK and FlgL from proteolysis and ensures that the substrates are efficiently transported to the export machinery (16, 17). To test if this was the case for the B. subtilis flgN deletion, the coding regions of flgK and flgL were integrated in the ΔflgN strain at a heterologous site on the chromosome under the control of an IPTG-inducible promoter. Induction of the flgK-flgL coding region was unable to restore motility to the ΔflgN strain, as determined by assaying swarming motility (Fig. 6A). To confirm that the flagellar filament was not polymerized upon overexpression of flgK-flgL, cellular protein samples (which include assembled flagella) were separated by SDS-PAGE and stained with Coomassie brilliant blue. Hag appears as a dominant protein band at ∼36 kDa (Fig. 6B) and can be easily identified by comparison with proteins harvested from the Δhag and wild-type strains (56). Moreover, the identity of the Hag protein was confirmed by mass spectrometry (see Fig. S1 in the supplemental material). Compared with the wild type, analysis of the cellular proteins for the ΔflgN strain indicated that Hag was not associated with the cell fraction (Fig. 6B). This is entirely consistent with the data presented above (Fig. 4 and 6A). As expected, the presence of the Hag band could be restored by the reintroduction of flgN on the chromosome upon induction with 50 μM IPTG (Fig. 6B). However, introduction of flgK-flgL in the ΔflgN background at a heterologous site could not complement the ΔflgN mutant with respect to Hag polymerization, even in the presence of 1 mM IPTG. The inability of flgK-flgL overexpression to compensate for deletion of flgN could be due to disruption of flagellar biosynthesis at an earlier stage. However, this possibility was ruled out as we demonstrated by Western blotting that the flagellar hook protein, FlgE, was detected in whole-cell lysates (which include assembled flagella) for both the wild-type and ΔflgN strains (see Fig. S2 in the supplemental material).
FIG 6.
Overexpression of flgK-flgL cannot complement a ΔflgN mutant. (A) Photographs of swarm expansion plates taken after 6 h of incubation at 37°C. Shown are the wild-type (3610), Δhag (DS1677), ΔflgN (NRS3570), and ΔflgN amyE::Phy-spank-flgK-flgL-lacI (NRS4043) strains without and with induction with 50 μM IPTG or 1 mM IPTG. (B) Coomassie gel analysis of cellular fractions of the 3610, Δhag (DS1677), ΔflgN (NRS3570), ΔflgN amyE::Phy-spank-flgN-lacI (NRS3578), and ΔflgN amyE::Phy-spank-flgK-flgL-lacI (NRS4043) strains without and with induction with 50 μM IPTG or 1 mM IPTG. The Hag protein was subsequently identified by mass spectrometry analysis and is marked with asterisks. MW, molecular weight in thousands.
The inability of heterologous flgK-flgL expression to complement the flgN mutant strain is suggestive of a strict dependence on FlgN for FlgK-FlgL protein stability or secretion in B. subtilis. In an attempt to test if FlgK was unstable in the absence of flgN, strains were constructed to enable detection of FlgK by fusing FlgK to a poly histidine epitope tag. However, the presence of such an epitope tag at either the N or C terminus of the protein rendered FlgK nonfunctional, as determined by a nonmotile phenotype (see Fig. S3A in the supplemental material). As a method of attempting to assess FlgK assembly at the flagellar hook junction, we aimed to purify flagellar hook-basal bodies from both the wild-type and ΔflgN strains and to analyze the protein components of the complex by mass spectrometry. While we were successfully able to enrich the flagellar fraction of the wild-type strain (see Fig. S3B and Table S4 in the supplemental material), we were unable to do so for the ΔflgN strain. This is most likely because this methodology is dependent on an intact flagellar filament for isolation of the complex, and the flgN mutant does not form a flagellar filament (Fig. 4). Finally, in S. Typhimurium, FlgN interacts directly with FlgK and FlgL to protect the proteins from proteolytic cleavage (25). To test if FlgN could interact with either FlgK or FlgL to perform a similar role in B. subtilis, bacterial two-hybrid experiments were undertaken. However, an interaction could not be detected (data not shown), which could be due to inactivity of the fusion protein. Thus, despite extensive efforts, we were unable to determine the stability of FlgK in the absence of flgN or if there was an interaction between the proteins.
In vivo analysis of the role of phosphorylation in controlling motility.
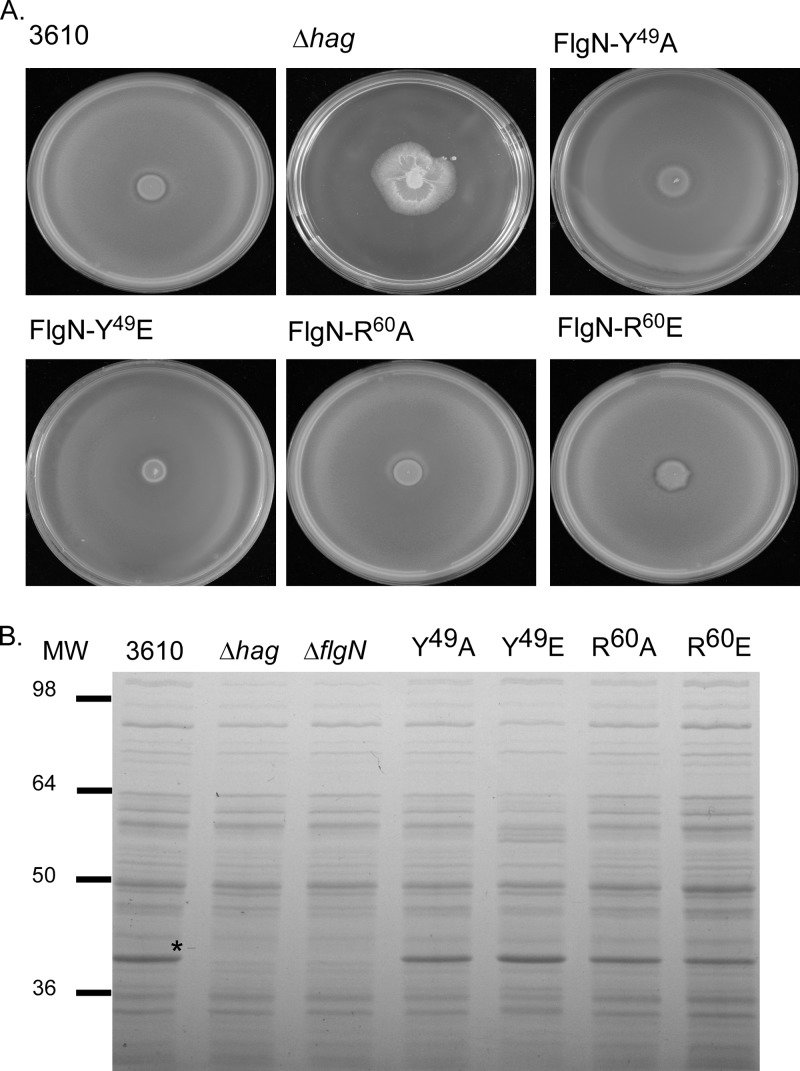
Global proteomic strategies have identified a plethora of targets for both tyrosine and arginine kinases in B. subtilis (28–30). One such target is FlgN, which can be tyrosine phosphorylated on amino acid 49 (29) and arginine phosphorylated on amino acid 60 (30). Having demonstrated that FlgN is required for flagellar biosynthesis, we were presented with the ideal system to assess the role of tyrosine and arginine phosphorylation in controlling protein function in vivo. Site-directed mutagenesis was used to mutate the chromosomal copy of flgN tyrosine 49 to alanine (Y49A) to assess the effects of preventing phosphorylation. As shown in Fig. 7, swarming motility was not affected. We next tested the impact of replacing tyrosine 49 with glutamic acid (Y49E) to mimic the negative charge associated with phosphorylation. Once more, the strain containing the mutant allele of FlgN (Y49E) exhibited a motility phenotype that was indistinguishable from that of the wild type (Fig. 7A). Subsequently, strains were constructed where the coding region of flgN on the chromosome was mutated such that the arginine at position 60 was replaced with either alanine or glutamic acid. The motility phenotypes of these strains were tested, and they were found to swarm at the same rate as the wild-type strain (Fig. 7A). SDS-PAGE analysis of whole-cell lysates also showed that Hag was synthesized and that the cellular localization was directly comparable to that of the wild type for each of the point mutation strains (Fig. 7B). These data demonstrate that mutation of the previously defined tyrosine or arginine phosphorylation site has no discernible impact on the function of FlgN in B. subtilis NCIB3610.
FIG 7.
Mutation of identified FlgN phosphorylation sites does not affect motility. (A) Photographs of swarm expansion plates taken after 6 h of incubation at 37°C. Shown are the wild-type (3610), Δhag (DS1677), flgN-Y49A (NRS3571), flgN-Y49E (NRS3724), flgN-R60A (NRS4063), and flgN-R60E (NRS4017) strains. (B) Coomassie gel analysis of cellular fractions of the 3610, Δhag (DS1677), flgN-Y49A (NRS3571), flgN-Y49E (NRS3724), flgN-R60A (NRS4063), and flgN-R60E (NRS4017) strains. The Hag protein is marked with asterisks. MW, molecular weight in thousands.
DISCUSSION
In this work we report that yvyG is required for the motility of B. subtilis. We demonstrate that the main role for YvyG is to enable flagellar filament polymerization. The data presented allow us to conclude that YvyG is indeed an orthologue of FlgN from S. Typhimurium, but in B. subtilis it would appear that there is a strict reliance on YvyG for the secretion and placement of FlgK and FlgL at the hook-filament junction. In light of these data, we suggest that YvyG be referred to as FlgN.
The role of FlgN in the regulation of flagellum biosynthesis.
This work suggests that in B. subtilis FlgN partially mediates flagellum biosynthesis through its ability to regulate hag transcription and translation. In wild-type B. subtilis the sigma factor σD (sigD) needed for hag transcription is transcribed only in a subpopulation of cells, resulting in bimodal expression of hag (11, 12). Deletion of flgN results is hag being transcribed in every cell, albeit at a lower level (Fig. 3), indicating that σD is active in every cell in this genetic background. Consistent with this, we did not observe any cell chaining in the absence of flgN, indicating that the σD-dependent autolysins (10) are also transcribed in all cells. It is known that σD can be regulated by transcription of the sigD gene and by interaction with the anti-sigma factor, FlgM (11). Therefore, the change in hag transcription observed upon deletion of flgN could be explained by a change in regulation by the anti-sigma factor, FlgM. In S. Typhimurium FlgM regulates the transcription of late-class σ28-regulated flagellar genes by both sequestering free σ28 and destabilizing the σ28 RNA polymerase holoenzyme complex (57). Upon completion of HBB assembly, FlgM is secreted, and σ28 is able to activate target promoters (58). It has been previously reported that FlgN is able to regulate the translation of FlgM in S. Typhimurium (59), therefore raising the possibility that in the B. subtilis ΔflgN strain translation of FlgM is decreased, allowing σD to trigger transcription of hag in all cells. The regulation of FlgM in B. subtilis is poorly understood, and so whether this is the case has yet to be determined.
Deletion of flgN in B. subtilis also results in a 2-fold decrease in hag translation (Fig. 4A). This effect could technically be due to translational regulation, as is seen for the ΔflgE strain (Fig. 4A) (55). Indeed, recent studies in B. subtilis have identified the RNA binding protein CsrA, the CsrA regulatory protein FliW, and the molecular chaperone FliS as having roles in controlling Hag translation or secretion (13, 55, 60). When cellular levels of Hag are depleted, FliW binds to CsrA, leaving it unable to occlude the hag Shine-Dalgarno sequence, allowing translation to proceed. However, when Hag protein accumulates in the cytoplasm, it is able to interact with and sequester FliW, resulting in CsrA-mediated repression of translation (55). Therefore, inhibition of translation by CsrA relies on accumulation of Hag within the cell. However, the data presented in Fig. 4B show that in the absence of flgN, Hag accumulates not in the cytoplasm but in the extracellular milieu. This not only suggests that CsrA is not responsible for the observed decrease in Hag translation but also is in keeping with the hypothesis that FlgN is required for the assembly of FlgK and FlgL; in the absence of the hook-filament junction, flagellin cannot be properly assembled and so accumulates in the extracellular milieu (53). For this reason we favor the hypothesis that the change in the transcriptional profile of hag is responsible for the decrease in translation observed.
The role of FlgN phosphorylation.
Tyrosine and arginine phosphorylation events have been implicated in the control of diverse biological processes in B. subtilis, including biofilm formation (38, 61), DNA replication (62), exopolysaccharide synthesis (63), the heat shock response (64), and potentially the regulation and/or assembly of the flagellar filament (30). The motility protein FlgN has been shown to be both tyrosine and arginine phosphorylated (29–31). Moreover, the subcellular localization of FlgN was reported to be impacted by deletion of the tyrosine kinase PtkA (31). However, site-directed mutagenesis of the reported FlgN tyrosine and arginine phosphorylation sites in vivo failed to impact the motility of B. subtilis (Fig. 7). These findings led us to conclude that a dominant role for phosphorylation of these residues does not exist. When these findings are considered in a wider context of the function of posttranslational modifications, they may not be surprising. For instance, in eukaryotes, it has been suggested that many phosphorylation events are nonfunctional or may occur at a very low stoichiometry such that they do not impact the function of the protein (65, 66). Indeed, two tyrosine kinases (61, 63) and one arginine kinase (30) have been identified in B. subtilis, but many more proteins have been identified as being phosphorylated, thereby suggesting that each kinase is promiscuous. This may imply that random encounters between kinases and phosphorylatable sites on different proteins might result in nonspecific and nonfunctional phosphorylation events. Alternatively, FlgN might act as a phosphate sink or store to remove free phosphate from the system (67).
Concluding remarks.
The regulation and biosynthesis of the bacterial flagellum are best understood for Gram-negative bacterial species such as S. Typhimurium. However, recent work on B. subtilis has begun to illuminate how Gram-positive bacterial species are able to coordinate flagellar assembly. These studies have uncovered key differences between Gram-negative and Gram-positive bacterial flagella as well as having highlighted many conserved mechanisms. The data presented here illustrate that FlgN from B. subtilis is essential for flagellar filament polymerization and therefore motility. We propose that Bs-FlgN is an orthologue of the S. Typhimurium chaperone protein FlgN and is required for the export and assembly of FlgK and FlgL at the hook-filament junction. By way of contrast, in a B. subtilis strain lacking flgN flagella are not detectable, and overexpression of flgK-flgL is not sufficient to overcome the motility defect exhibited by the ΔflgN strain. Therefore, while Bs-FlgN bears great functional similarity to ST-FlgN, there are crucial differences which suggest that there is a stricter dependence on FlgN for the export of FlgK and FlgL in B. subtilis. Overall, this work further emphasizes the previously underappreciated differences in flagellar gene regulation between Gram-positive and Gram-negative bacterial species.
Supplementary Material
ACKNOWLEDGMENTS
L.S.C. is the recipient of a Wellcome Trust Ph.D. studentship (093714/Z/10/Z). V.L.M. was funded by a Biotechnology and Biological Sciences Research Council grant (BB/I006915/1). A.O. was the recipient of a Biotechnology and Biological Sciences Research Council Doctoral Training Account grant (BB/D526161/1). The microscopy facilities are supported by a Wellcome Trust Strategic Award grant (083524/Z/07/Z).
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank Kürsad Turgay for the kind gift of the anti-Hag antibody, Daniel Kearns for the kind gift of the anti-FlgE antibody and helpful discussions, Rosemary Clarke for assistance with flow cytometry, FingerPrints Proteomics and Mass Spectrometry Facility, University of Dundee, and Shin-Ichi Aizawa for his helpful (and repeated) advice regarding the purification of flagellar hook-basal bodies.
Footnotes
Published ahead of print 4 April 2014
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.01599-14.
REFERENCES
- 1.Chevance FF, Hughes KT. 2008. Coordinating assembly of a bacterial macromolecular machine. Nat. Rev. Microbiol. 6:455–465. 10.1038/nrmicro1887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Toole GA, Kaplan HB, Kolter R. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49–79. 10.1146/annurev.micro.54.1.49 [DOI] [PubMed] [Google Scholar]
- 3.Cairns LS, Marlow VL, Bissett E, Ostrowski A, Stanley-Wall NR. 2013. A mechanical signal transmitted by the flagellum controls signalling in Bacillus subtilis. Mol. Microbiol. 90:6–21. 10.1111/mmi.12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serra DO, Richter AM, Klauck G, Mika F, Hengge R. 2013. Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio. 4:e00103–13. 10.1128/mBio.00103-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marquez-Magana LM, Chamberlin MJ. 1994. Characterization of the sigD transcription unit of Bacillus subtilis. J. Bacteriol. 176:2427–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.West JT, Estacio W, Marquez-Magana L. 2000. Relative roles of the fla/che PA, PD-3, and PsigD promoters in regulating motility and sigD expression in Bacillus subtilis. J. Bacteriol. 182:4841–4848. 10.1128/JB.182.17.4841-4848.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helmann JD, Marquez LM, Chamberlin MJ. 1988. Cloning, sequencing, and disruption of the Bacillus subtilis sigma 28 gene. J. Bacteriol. 170:1568–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen YF, Helmann JD. 1992. Restoration of motility to an Escherichia coli fliA flagellar mutant by a Bacillus subtilis sigma factor. Proc. Natl. Acad. Sci. U. S. A. 89:5123–5127. 10.1073/pnas.89.11.5123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mirel DB, Lauer P, Chamberlin MJ. 1994. Identification of flagellar synthesis regulatory and structural genes in a sigma D-dependent operon of Bacillus subtilis. J. Bacteriol. 176:4492–4500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Marquez LM, Helmann JD, Ferrari E, Parker HM, Ordal GW, Chamberlin MJ. 1990. Studies of sigma D-dependent functions in Bacillus subtilis. J. Bacteriol. 172:3435–3443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cozy LM, Kearns DB. 2010. Gene position in a long operon governs motility development in Bacillus subtilis. Mol. Microbiol. 76:273–285. 10.1111/j.1365-2958.2010.07112.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kearns DB, Losick R. 2005. Cell population heterogeneity during growth of Bacillus subtilis. Genes Dev. 19:3083–3094. 10.1101/gad.1373905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mukherjee S, Babitzke P, Kearns DB. 2013. FliW and FliS function independently to control cytoplasmic flagellin levels in Bacillus subtilis. J. Bacteriol. 195:297–306. 10.1128/JB.01654-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Titz B, Rajagopala SV, Ester C, Hauser R, Uetz P. 2006. Novel conserved assembly factor of the bacterial flagellum. J. Bacteriol. 188:7700–7706. 10.1128/JB.00820-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bennett JC, Hughes C. 2000. From flagellum assembly to virulence: the extended family of type III export chaperones. Trends Microbiol. 8:202–204. 10.1016/S0966-842X(00)01751-0 [DOI] [PubMed] [Google Scholar]
- 16.Aldridge P, Karlinsey J, Hughes KT. 2003. The type III secretion chaperone FlgN regulates flagellar assembly via a negative feedback loop containing its chaperone substrates FlgK and FlgL. Mol. Microbiol. 49:1333–1345. 10.1046/j.1365-2958.2003.03637.x [DOI] [PubMed] [Google Scholar]
- 17.Thomas J, Stafford GP, Hughes C. 2004. Docking of cytosolic chaperone-substrate complexes at the membrane ATPase during flagellar type III protein export. Proc. Natl. Acad. Sci. U. S. A. 101:3945–3950. 10.1073/pnas.0307223101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Evans LD, Stafford GP, Ahmed S, Fraser GM, Hughes C. 2006. An escort mechanism for cycling of export chaperones during flagellum assembly. Proc. Natl. Acad. Sci. U. S. A. 103:17474–17479. 10.1073/pnas.0605197103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minamino T, MacNab RM. 2000. Interactions among components of the Salmonella flagellar export apparatus and its substrates. Mol. Microbiol. 35:1052–1064. 10.1046/j.1365-2958.2000.01771.x [DOI] [PubMed] [Google Scholar]
- 20.Minamino T, Shimada M, Okabe M, Saijo-Hamano Y, Imada K, Kihara M, Namba K. 2010. Role of the C-terminal cytoplasmic domain of FlhA in bacterial flagellar type III protein export. J. Bacteriol. 192:1929–1936. 10.1128/JB.01328-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bange G, Kummerer N, Engel C, Bozkurt G, Wild K, Sinning I. 2010. FlhA provides the adaptor for coordinated delivery of late flagella building blocks to the type III secretion system. Proc. Natl. Acad. Sci. U. S. A. 107:11295–11300. 10.1073/pnas.1001383107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Minamino T, Namba K. 2008. Distinct roles of the FliI ATPase and proton motive force in bacterial flagellar protein export. Nature 451:485–488. 10.1038/nature06449 [DOI] [PubMed] [Google Scholar]
- 23.Paul K, Erhardt M, Hirano T, Blair DF, Hughes KT. 2008. Energy source of flagellar type III secretion. Nature 451:489–492. 10.1038/nature06497 [DOI] [PubMed] [Google Scholar]
- 24.Ozin AJ, Claret L, Auvray F, Hughes C. 2003. The FliS chaperone selectively binds the disordered flagellin C-terminal D0 domain central to polymerisation. FEMS Microbiol. Lett. 219:219–224. 10.1016/S0378-1097(02)01208-9 [DOI] [PubMed] [Google Scholar]
- 25.Bennett JC, Thomas J, Fraser GM, Hughes C. 2001. Substrate complexes and domain organization of the Salmonella flagellar export chaperones FlgN and FliT. Mol. Microbiol. 39:781–791. 10.1046/j.1365-2958.2001.02268.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser GM, Bennett JC, Hughes C. 1999. Substrate-specific binding of hook-associated proteins by FlgN and FliT, putative chaperones for flagellum assembly. Mol. Microbiol. 32:569–580. 10.1046/j.1365-2958.1999.01372.x [DOI] [PubMed] [Google Scholar]
- 27.Pallen MJ, Penn CW, Chaudhuri RR. 2005. Bacterial flagellar diversity in the post-genomic era. Trends Microbiol. 13:143–149. 10.1016/j.tim.2005.02.008 [DOI] [PubMed] [Google Scholar]
- 28.Levine A, Vannier F, Absalon C, Kuhn L, Jackson P, Scrivener E, Labas V, Vinh J, Courtney P, Garin J, Seror SJ. 2006. Analysis of the dynamic Bacillus subtilis Ser/Thr/Tyr phosphoproteome implicated in a wide variety of cellular processes. Proteomics 6:2157–2173. 10.1002/pmic.200500352 [DOI] [PubMed] [Google Scholar]
- 29.Macek B, Mijakovic I, Olsen JV, Gnad F, Kumar C, Jensen PR, Mann M. 2007. The serine/threonine/tyrosine phosphoproteome of the model bacterium Bacillus subtilis. Mol. Cell. Proteomics 6:697–707. 10.1074/mcp.M600464-MCP200 [DOI] [PubMed] [Google Scholar]
- 30.Elsholz AK, Turgay K, Michalik S, Hessling B, Gronau K, Oertel D, Mader U, Bernhardt J, Becher D, Hecker M, Gerth U. 2012. Global impact of protein arginine phosphorylation on the physiology of Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 109:7451–7456. 10.1073/pnas.1117483109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jers C, Pedersen MM, Paspaliari DK, Schutz W, Johnsson C, Soufi B, Macek B, Jensen PR, Mijakovic I. 2010. Bacillus subtilis BY-kinase PtkA controls enzyme activity and localization of its protein substrates. Mol. Microbiol. 77:287–299. 10.1111/j.1365-2958.2010.07227.x [DOI] [PubMed] [Google Scholar]
- 32.Garnak M, Reeves HC. 1979. Phosphorylation of Isocitrate dehydrogenase of Escherichia coli. Science 203:1111–1112. 10.1126/science.34215 [DOI] [PubMed] [Google Scholar]
- 33.Deutscher J, Kuster E, Bergstedt U, Charrier V, Hillen W. 1995. Protein kinase-dependent HPr-CcpA interaction links glycolytic activity to carbon catabolite repression in Gram-positive bacteria. Mol. Microbiol. 15:1049–1053. 10.1111/j.1365-2958.1995.tb02280.x [DOI] [PubMed] [Google Scholar]
- 34.Cozy LM, Phillips AM, Calvo RA, Bate AR, Hsueh YH, Bonneau R, Eichenberger P, Kearns DB. 2012. SlrA/SinR/SlrR inhibits motility gene expression upstream of a hypersensitive and hysteretic switch at the level of sD in Bacillus subtilis. Mol. Microbiol. 83:1210–1228. 10.1111/j.1365-2958.2012.08003.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hsueh YH, Cozy LM, Sham LT, Calvo RA, Gutu AD, Winkler ME, Kearns DB. 2011. DegU-phosphate activates expression of the anti-sigma factor FlgM in Bacillus subtilis. Mol. Microbiol. 81:1092–1108. 10.1111/j.1365-2958.2011.07755.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kutsukake K, Okada T, Yokoseki T, Iino T. 1994. Sequence analysis of the flgA gene and its adjacent region in Salmonella Typhimurium, and identification of another flagellar gene, flgN. Gene 143:49–54. 10.1016/0378-1119(94)90603-3 [DOI] [PubMed] [Google Scholar]
- 37.Verhamme DT, Kiley TB, Stanley-Wall NR. 2007. DegU co-ordinates multicellular behaviour exhibited by Bacillus subtilis. Mol. Microbiol. 65:554–568. 10.1111/j.1365-2958.2007.05810.x [DOI] [PubMed] [Google Scholar]
- 38.Kiley TB, Stanley-Wall NR. 2010. Post-translational control of Bacillus subtilis biofilm formation mediated by tyrosine phosphorylation. Mol. Microbiol. 78:947–963. 10.1111/j.1365-2958.2010.07382.x [DOI] [PubMed] [Google Scholar]
- 39.Arnaud M, Chastanet A, Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891. 10.1128/AEM.70.11.6887-6891.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu H, Naismith JH. 2009. A simple and efficient expression and purification system using two newly constructed vectors. Protein Expr. Purif. 63:102–111. 10.1016/j.pep.2008.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ostrowski A, Mehert A, Prescott A, Kiley TB, Stanley-Wall NR. 2011. YuaB functions synergistically with the exopolysaccharide and TasA amyloid fibers to allow biofilm formation by Bacillus subtilis. J. Bacteriol. 193:4821–4831. 10.1128/JB.00223-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. 10.1038/msb.2011.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 292:195–202. 10.1006/jmbi.1999.3091 [DOI] [PubMed] [Google Scholar]
- 44.Buchan DW, Minneci F, Nugent TC, Bryson K, Jones DT. 2013. Scalable web services for the PSIPRED protein Analysis Workbench. Nucleic Acids Res. 41:W349–357. 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blair KM, Turner L, Winkelman JT, Berg HC, Kearns DB. 2008. A molecular clutch disables flagella in the Bacillus subtilis biofilm. Science 320:1636–1638. 10.1126/science.1157877 [DOI] [PubMed] [Google Scholar]
- 46.Allan C, Burel JM, Moore J, Blackburn C, Linkert M, Loynton S, Macdonald D, Moore WJ, Neves C, Patterson A, Porter M, Tarkowska A, Loranger B, Avondo J, Lagerstedt I, Lianas L, Leo S, Hands K, Hay RT, Patwardhan A, Best C, Kleywegt GJ, Zanetti G, Swedlow JR. 2012. OMERO: flexible, model-driven data management for experimental biology. Nat. Methods 9:245–253. 10.1038/nmeth.1896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Murray EJ, Strauch MA, Stanley-Wall NR. 2009. σX is involved in controlling Bacillus subtilis biofilm architecture through the AbrB homologue Abh. J. Bacteriol. 191:6822–6832. 10.1128/JB.00618-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Karlinsey JE, Tanaka S, Bettenworth V, Yamaguchi S, Boos W, Aizawa SI, Hughes KT. 2000. Completion of the hook-basal body complex of the Salmonella Typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 37:1220–1231. 10.1046/j.1365-2958.2000.02081.x [DOI] [PubMed] [Google Scholar]
- 49.Murray EJ, Kiley TB, Stanley-Wall NR. 2009. A pivotal role for the response regulator DegU in controlling multicellular behaviour. Microbiology 155:1–8. 10.1099/mic.0.023903-0 [DOI] [PubMed] [Google Scholar]
- 50.Miller J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 51.Aizawa SI, Dean GE, Jones CJ, Macnab RM, Yamaguchi S. 1985. Purification and characterization of the flagellar hook-basal body complex of Salmonella Typhimurium. J. Bacteriol. 161:836–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kubori T, Okumura M, Kobayashi N, Nakamura D, Iwakura M, Aizawa SI. 1997. Purification and characterization of the flagellar hook-basal body complex of Bacillus subtilis. Mol. Microbiol. 24:399–410. 10.1046/j.1365-2958.1997.3341714.x [DOI] [PubMed] [Google Scholar]
- 53.Gygi D, Fraser G, Dufour A, Hughes C. 1997. A motile but non-swarming mutant of Proteus mirabilis lacks FlgN, a facilitator of flagella filament assembly. Mol. Microbiol. 25:597–604. 10.1046/j.1365-2958.1997.5021862.x [DOI] [PubMed] [Google Scholar]
- 54.Fraser GM, Hughes C. 1999. Swarming motility. Curr. Opin. Microbiol. 2:630–635. 10.1016/S1369-5274(99)00033-8 [DOI] [PubMed] [Google Scholar]
- 55.Mukherjee S, Yakhnin H, Kysela D, Sokoloski J, Babitzke P, Kearns DB. 2011. CsrA-FliW interaction governs flagellin homeostasis and a checkpoint on flagellar morphogenesis in Bacillus subtilis. Mol. Microbiol. 82:447–461. 10.1111/j.1365-2958.2011.07822.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Diethmaier C, Pietack N, Gunka K, Wrede C, Lehnik-Habrink M, Herzberg C, Hubner S, Stulke J. 2011. A novel factor controlling bistability in Bacillus subtilis: the YmdB protein affects flagellin expression and biofilm formation. J. Bacteriol. 193:5997–6007. 10.1128/JB.05360-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chadsey MS, Karlinsey JE, Hughes KT. 1998. The flagellar anti-sigma factor FlgM actively dissociates Salmonella Typhimurium sigma28 RNA polymerase holoenzyme. Genes Dev. 12:3123–3136. 10.1101/gad.12.19.3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kutsukake K. 1994. Excretion of the anti-sigma factor through a flagellar substructure couples flagellar gene expression with flagellar assembly in Salmonella Typhimurium. Mol. Gen. Genet. 243:605–612 [DOI] [PubMed] [Google Scholar]
- 59.Karlinsey JE, Lonner J, Brown KL, Hughes KT. 2000. Translation/secretion coupling by type III secretion systems. Cell 102:487–497. 10.1016/S0092-8674(00)00053-2 [DOI] [PubMed] [Google Scholar]
- 60.Yakhnin H, Pandit P, Petty TJ, Baker CS, Romeo T, Babitzke P. 2007. CsrA of Bacillus subtilis regulates translation initiation of the gene encoding the flagellin protein (hag) by blocking ribosome binding. Mol. Microbiol. 64:1605–1620. 10.1111/j.1365-2958.2007.05765.x [DOI] [PubMed] [Google Scholar]
- 61.Gerwig J, Kiley TB, Gunka K, Stanley-Wall N, Stulke J. 2014. The protein tyrosine kinases EpsB and PtkA differentially affect biofilm formation in Bacillus subtilis. Microbiology 160:682–691. 10.1099/mic.0.074971-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Petranovic D, Michelsen O, Zahradka K, Silva C, Petranovic M, Jensen PR, Mijakovic I. 2007. Bacillus subtilis strain deficient for the protein-tyrosine kinase PtkA exhibits impaired DNA replication. Mol. Microbiol. 63:1797–1805. 10.1111/j.1365-2958.2007.05625.x [DOI] [PubMed] [Google Scholar]
- 63.Mijakovic I, Poncet S, Boel G, Maze A, Gillet S, Jamet E, Decottignies P, Grangeasse C, Doublet P, Le Marechal P, Deutscher J. 2003. Transmembrane modulator-dependent bacterial tyrosine kinase activates UDP-glucose dehydrogenases. EMBO J. 22:4709–4718. 10.1093/emboj/cdg458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fuhrmann J, Schmidt A, Spiess S, Lehner A, Turgay K, Mechtler K, Charpentier E, Clausen T. 2009. McsB is a protein arginine kinase that phosphorylates and inhibits the heat-shock regulator CtsR. Science 324:1323–1327. 10.1126/science.1170088 [DOI] [PubMed] [Google Scholar]
- 65.Landry CR, Levy ED, Michnick SW. 2009. Weak functional constraints on phosphoproteomes. Trends Genet. 25:193–197. 10.1016/j.tig.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 66.Beltrao P, Albanese V, Kenner LR, Swaney DL, Burlingame A, Villen J, Lim WA, Fraser JS, Frydman J, Krogan NJ. 2012. Systematic functional prioritization of protein posttranslational modifications. Cell 150:413–425. 10.1016/j.cell.2012.05.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sourjik V, Schmitt R. 1998. Phosphotransfer between CheA, CheY1, and CheY2 in the chemotaxis signal transduction chain of Rhizobium meliloti. Biochemistry 37:2327–2335. 10.1021/bi972330a [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.